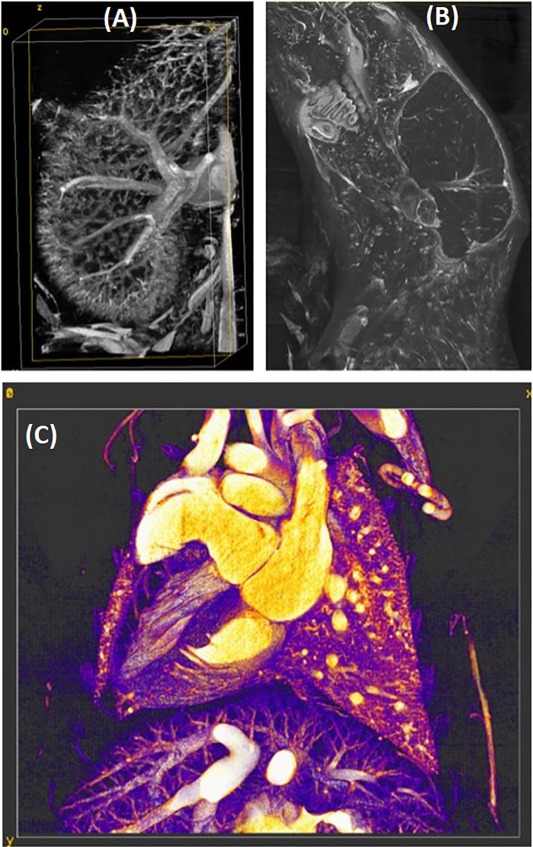
Our team is part of the Radiology Department and its Brain Imaging and Analysis Center, as well as part of the Neurology Department. At the center of our methods, we use MRI, which is particularly suitable for brain imaging, and capable of providing multiple imaging contrasts and quantitative biomarkers including those based on morphometry, relaxation parameters, quantitative susceptibility or diffusion, from which we derive tractography. Diffusion weighted imaging is an important tool for studying brain microstructure, and the connectivity amongst gray matter regions.
Our work involves image segmentation, morphometry and shape analysis, as well as in integrating information from MRI with genetics, and behavior. Our approaches target: 1) phenotyping the neuroanatomy using imaging; 2) uncovering the link between structural and functional changes, the genetic bases, and environmental factors. Our results consist of methods and tools for comprehensive phenotyping.
We aim to integrate imaging data from multiple modalities, and across scales. We use high-performance cluster computing to accelerate our image analysis. We use compressed sensing image reconstruction, and process large image arrays using deformable registration, perform segmentation based on multiple image contrasts including diffusion tensor imaging, as well as voxel based analysis, and graph analysis for connectomics.
We have developed high resolution, multivariate population atlases for animal models. At BIAC we focus on developing multivariate biomarkers and identifying vulnerable networks based on genetic risk for Alzheimer’s disease.
We continue working on multivariate image analysis and predictive modeling approaches to help better understand early biomarkers for human disease indirectly through mouse models, as well as directly. The hope is to develop strategies that will help increase the rate at which we grow our current understanding of gray and white matter changes in neurological and psychiatric conditions.
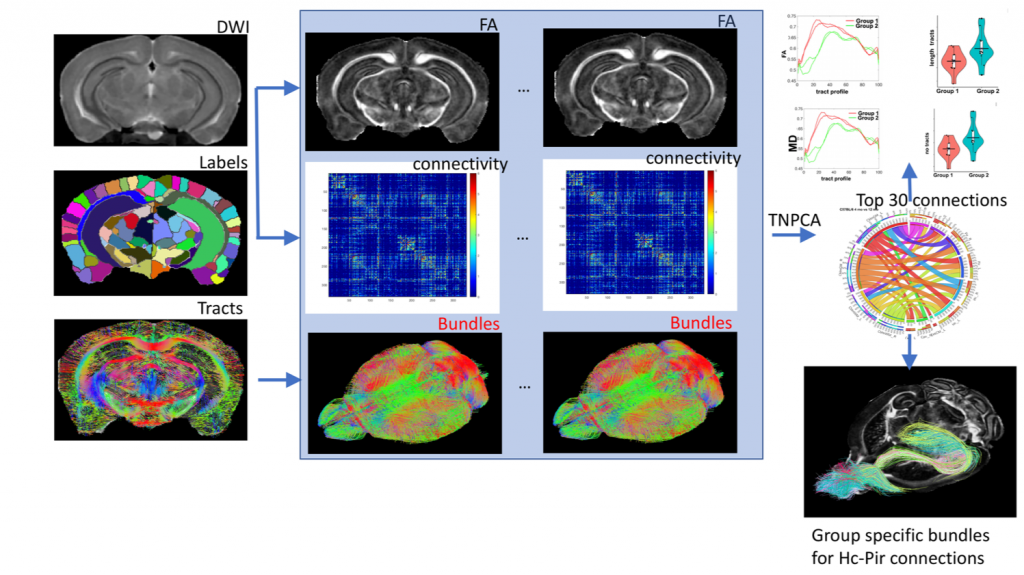
Our workflows incorporate atlas based segmentation, using a minimum deformation template, generated specifically to a particular study. We derive connectomes to identify vulnerable networks, and then analyze the properties of these connections. Read more about our work in this open access publication.