(M)ulti-(C)hannel CT (R)econstruction Toolkit
An open-source toolkit for advanced applications of preclinical and clinical x-ray CT.
Copyright (C) 2022-2023, Quantitative Imaging and Analysis Lab, Duke University
A beta release of version 2.0 of the MCR Toolkit for Python is now available!
About
The MCR Toolkit has evolved over nearly a decade, first to address unique challenges associated with in vivo preclinical micro-CT, and now to support translation of advances in CT reconstruction between the preclinical and clinical domains. As the name suggests, the Toolkit specializes in solving “multi-channel” reconstruction problems: multi-energy (ME) CT (dual-energy, photon-counting), dynamic time-resolved (TR) CT (cardiac, perfusion), and combined multi-energy and time-resolved (METR) CT. The Toolkit strives to balance the computational demands of multi-channel reconstruction problems against the need for numerically robust denoising and reconstruction operators required to solve real-world problems. Currently, the Toolkit is mainly developed for MATLAB and Linux; however, support for Windows operating systems is maintained. A Python interface also exists to support deep learning projects.
Contact Information:
The initial public release of the Toolkit was developed by:
Darin P. Clark, PhD
Asst. Professor in Radiology
Duke University Medical Center
dpc18@duke.edu
Quantitative Imaging and Analysis Lab
Center for Virtual Imaging Trials
If you are interested in contributing to the Toolkit’s development, please do not hesitate to contact me.
Contributions:
We acknowledge Samuel Johnston, PhD, for his work on early versions of the MCR Toolkit.
Toolkit Features:
- Rapid prototyping of reconstruction pipelines through MATLAB
- Python support for interoperability with deep learning packages (in development)
- Unified iterative reconstruction functions for single energy (SE), ME, TR, and METR reconstruction problems.
- Matched projection and backprojection operators supporting flat and cylindrical CT detectors and multiple GPUs
- Data adaptation: semi-automatic and automatic adjustment of regularization parameters
- GPU-based denoising and regularization algorithms:
- Rank-Sparse Kernel Regression (RSKR): iterative application of BF to weighted singular vectors to approximately homogenize noise variance across ME CT data sets.
- Patch-based Singular Value Thresholding (pSVT): singular value thresholding applied to patches extracted along the time dimension to improve denoising performance.
- Dictionary sparse coding: patch-wise coding using a learned dictionary of atoms; a precursor to convolutional sparse coding and convolutional neural networks
Citation
Continued development of the MCR Toolkit relies on research grants. If you use the MCR Toolkit in your work, please cite the following publication:
Clark DP, Badea CT. MCR toolkit: A GPU-based toolkit for multi-channel reconstruction of preclinical and clinical x-ray CT data. Med. Phys.. 2023;1-22. https://doi.org/10.1002/mp.16532
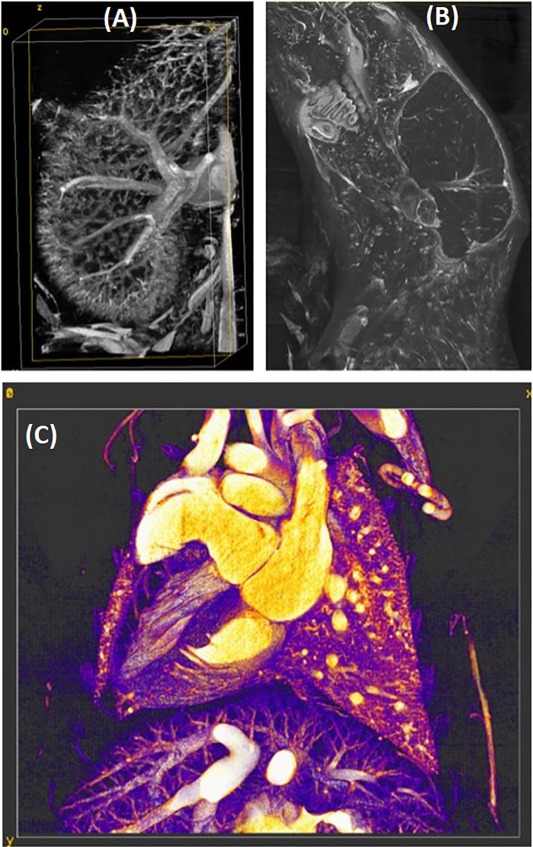
Gallery

Photon-counting CT imaging of a sarcoma tumor on the flank of a mouse, three days after the injection of iodine liposomes. Maximum intensity projections are shown in axial (left) and sagittal (right) orientations. Post-reconstruction material decomposition was applied to decompose the resulting multi-energy CT data into water (gray), iodine (red), and calcium (green) maps. Collaborators: David Kirsch, MD, PhD; Chang-Lung Lee, PhD; Yvonne M. Mowery, MD, PhD.


Photon-counting CT imaging of the heart in a mouse model of atherosclerosis (ApoE knockout). Maximum intensity projections are shown through the heart following time and energy resolved reconstruction and post-reconstruction material decomposition: water (gray), iodine (red), calcium and gold (green). Collaborators: Chang-Lung Lee, PhD; Matthew Holbrook, PhD.

Photon-counting CT perfusion imaging of a mouse model of head and neck cancer. A 2D coronal slice is shown after time and energy resolved reconstruction and post-reconstruction material decomposition: water (gray), iodine (red), calcium (green). Collaborators: Yvonne M. Mowery, MD, PhD; Alex Allphin.
System Requirements
- At least one NVIDIA GPU with 16 GB of GPU memory and support for compute capability 6.1 (Pascal) or higher. When using multiple GPUs, the Toolkit currently expects all GPUs used to be similar.
- NVIDIA CUDA Toolkit 9.0 or newer (https://developer.nvidia.com/cuda-toolkit)
- MATLAB version 2018a or newer with compatible C/C++ MEX compiler (e.g., gcc for Linux, Visual C++ for Windows)
- Linux operating system with NVIDIA driver support (highly recommended)
-OR-
Windows operating system - 128 GB of system RAM (TR and METR iterative reconstruction)
- Currently, the Toolkit relies on the Tools for NIfTI and ANALYZE image package for MATLAB to save and load NIfTI image volume files
Installation Instructions
MATLAB Interface
- Download or clone a copy of the repository from GitLab (Link).
- In MATLAB, open the master_builder.m script from the Toolkit root directory.
- In the first active cell of the master_builder.m script, set the following absolute paths for your system and operating system:
- mex_includes: absolute path to the MATLAB include subdirectory
- toolkit_path: absolute path to the MCR Toolkit root directory
- cuda_libraries: absolute path to the CUDA lib subdirectory
- cuda_includes: absolute path to the CUDA include subdirectory
- c_compiler: absolute path to your C/C++ compiler
- mex_path: absolute path to MATLAB’s mex subdirectory
- nifti_toolkit: absolute path to the Tools for NIfTI and ANALYZE image directory
- Run the entire master_builder.m script to compile and test the components of the Toolkit and to properly set up your MATLAB paths for using the Toolkit.
MATLAB Interface – Windows
For performance reasons and multi-GPU computing, Linux is strongly recommended; however, the Toolkit can also be run on a single GPU from Windows through MATLAB. After following the set-up above, follow these additional instructions.
To get MALTAB to set the appropriate environment variables automatically when MATLAB starts up, follow these instructions.
1. Execute the following from the MATLAB command window: edit(fullfile(userpath,’startup.m’))
2. Append the following to the startup.m file, assuming the GPU with index 0 is the desired GPU to use for computation: setenv(‘CUDA_MANAGED_FORCE_DEVICE_ALLOC’,’1′); setenv(‘CUDA_VISIBLE_DEVICES’,num2str(0));
3. Restart MATLAB
NOTE: Computing performance under Windows may further improve using NVIDIA’s TCC driver option. See here for details.
Python Interface (Beta)
- NOTE: Python support and deep learning integration are currently under development. Python support has not been tested under Windows and version and system requirements are still undergoing evaluation.
- Edit the Makefile in the DD_operators_vXX subdirectory of the Toolkit.
- CC: absolute path to C compiler.
- CUDA_LIBS2: ‘-L’ + absolute path to ‘targets/x86_64-linux/lib/’ CUDA subdirectory
- CUDA_I: ‘-I’ + absolute path to ‘targets/x86_64-linux/include/’ CUDA subdirectory
- Run ‘make –always-make’ from within the DD_operators_vXX subdirectory of the Toolkit.
- Confirm that the “DD_operators_vXX.so” library has been built successfully in the DD_operators_vXX subdirectory of the Toolkit.
- Consult the ‘/Demo_Scripts/Recon_Scripts/Python_InterfaceSample_3D_Recon_w_Python_v1.py’ demo for an example of using the Python interface for reconstruction.
- Before running, update the toolkit_path and recon_lib_path variables to point to the appropriate directories. Also, import the Python file ‘Python/DD_Recon.py’ into your Python project.
Denoising and Reconstruction Demos
- Supplemental data required to run the MCR Toolkit demo scripts can be found here.
- For convenience, demo scripts can be run from the run_demos.m script in the Toolkit root directory after completing a MATLAB installation. Note that some demos are commented out due to restrictions on sharing supplemental data; however, complete scripts are still provided in the Demo_Scripts subdirectory of the Toolkit.
- Demo_Scripts/Denoising_Scripts: Preclinical and clinical examples of denoising METR CT data with RSKR and pSVT. The clinical example also performs post reconstruction material decomposition.
- Demo_Scripts/DukeSim_Recon_Scripts: Cardiac CT reconstruction of XCAT phantom projection data simulated using DukeSim.
- Demo_Scripts/Recon_Scripts/MOBY_Phantom_METR: SE, ME, TR, and METR reconstruction of MOBY phantom projection data with analytical and iterative reconstruction.
- Demo_Scripts/Recon_Scripts/Preclinical_CT_METR: A complete reconstruction pipeline for in vivo, cardiac, photon-counting x-ray CT data acquired in a mouse model of atherosclerosis. The photon-counting detector was a SANTIS 0804 ME prototype detector on loan from DECTRIS, AG. The detector has a 1 mm thick CdTe sensor, 150-μm isotropic pixels (515×257), and four independent and fully adjustable energy thresholds (set to 25, 34, 40, and 55 keV).
- Demo_Scripts/Recon_Scripts/Siemens_Flash_DS_DE: Dual-source, dual-energy CT reconstruction of Multi-Energy CT Phantom projection data acquired with a Siemens Flash scanner.
Funding and Support
Funding for this work was provided by the National Institutes of Health: National Institute on Aging (RF1AG070149, supplement RF1AG070149-01S1), National Cancer Institute (U24CA220245), and the National Institute of Biomedical Imaging and Bioengineering (R01EB001838, P41EB028744).
The MCR Toolkit has been developed to support collaborative research projects with Siemens Healthineers, AG, and DECTRIS, AG; however, neither company has provided direct support for the Toolkit’s development.
References
Matched projection and backprojection operators
Long, Y., Fessler, J. A., & Balter, J. M. (2010).
3D forward and back-projection for X-ray CT using separable footprints.
IEEE transactions on medical imaging, 29(11), 1839-1850.
Approximate, analytical reconstruction of helical CT data
Stierstorfer, K., Rauscher, A., Boese, J., Bruder, H., Schaller, S., & Flohr, T. (2004).
Weighted FBP—a simple approximate 3D FBP algorithm for multislice spiral CT with good dose usage for arbitrary pitch.
Physics in Medicine & Biology, 49(11), 2209.
Projection matrices for efficient geometry calculations with flat detectors
Galigekere, R. R., Wiesent, K., & Holdsworth, D. W. (2003).
Cone-beam reprojection using projection-matrices.
IEEE transactions on medical imaging, 22(10), 1202-1214.
BiCGSTAB(l) linear solver (implemented in the MCR Toolkit)
Sleijpen, G. L., & Van Gijzen, M. B. (2010).
Exploiting BiCGstab (ℓ) strategies to induce dimension reduction.
SIAM journal on scientific computing, 32(5), 2687-2709.
Algorithm 3.1
Denoising with patch-based singular value thresholding (pSVT, implemented in the MCR Toolkit)
Kim, K., Ye, J. C., Worstell, W., Ouyang, J., Rakvongthai, Y., El Fakhri, G., & Li, Q. (2014).
Sparse-view spectral CT reconstruction using spectral patch-based low-rank penalty.
IEEE transactions on medical imaging, 34(3), 748-760.
Denoising with rank-sparse kernel regression (RSKR, implemented in the MCR Toolkit)
Clark, D. P., & Badea, C. T. (2017).
Hybrid spectral CT reconstruction.
PloS one, 12(7), e0180324.
Denoising with dictionary sparse coding (implemented in the MCR Toolkit)
Rubinstein, R., Zibulevsky, M., & Elad, M. (2008). Efficient implementation of the K-SVD algorithm using batch orthogonal matching pursuit (No. CS Technion report CS-2008-08). Computer Science Department, Technion.
DukeSim CT Simulator
Abadi, E., Harrawood, B., Sharma, S., Kapadia, A., Segars, W. P., & Samei, E. (2018).
DukeSim: a realistic, rapid, and scanner-specific simulation framework in computed tomography.
IEEE transactions on medical imaging, 38(6), 1457-1465.
MOBY mouse phantom
Segars, W. P., Tsui, B. M., Frey, E. C., Johnson, G. A., & Berr, S. S. (2004).
Development of a 4-D digital mouse phantom for molecular imaging research.
Molecular Imaging & Biology, 6(3), 149-159.
XCAT human phantom
W. Segars, G. Sturgeon, S. Mendonca, J. Grimes, and B. M. Tsui. (2010)
4D XCAT phantom for multimodality imaging research.
Medical Physics 37(9), 4902-4915.
Licensing
Copyright (C) 2022-2023, Quantitative Imaging and Analysis Lab, Duke University
This program is free software: you can redistribute it and/or modify
it under the terms of the GNU General Public License as published by
the Free Software Foundation, either version 3 of the License, or
(at your option) any later version.
This program is distributed in the hope that it will be useful,
but WITHOUT ANY WARRANTY; without even the implied warranty of
MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
GNU General Public License for more details.
You should have received a copy of the GNU General Public License
along with this program. If not, see <https://www.gnu.org/licenses/>.