Key:
# stands for contributions
* stands for co-correspondence
Google Scholar Publications
Search Pub Med
PUBLICATIONS

127. REV7 associates with ATRIP and inhibits ATR kinase activity. Biller M, Kabir S, Nipper S, Allen S, Kayali Y, Kuncik S, Sasanuma H, Zhou P, Vaziri C, Tomida J. Nucleic Acids Research 2026 (accepted). (doi: NAR-00314-Q-2025.R2)

126. REV1 Loss Triggers a G2/M Cell-Cycle Arrest Through Dysregulation of Mitotic Regulators. Buntin B, Guyette M, Gupta V, Ikeh K, Bhattacharya S, Lamkin EN, Lafuze A, Rio-Guerra R, Hong J, Zhou P, Chatterjee N. Genes 2026; 17: 44. (doi: 10.3390/genes17010044)
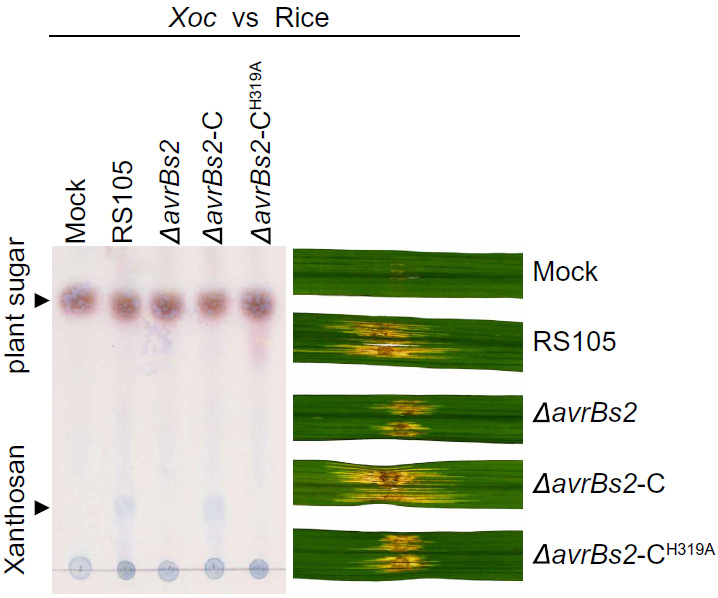
125. A bacterial nutrition strategy for plant disease control. Wang S, Zhu L, Tian M, Wu W, Hu X, Li X, Wang J, Zhu Y, Xu J, Mou B, Yang J, Cui F, Li D, Cheng J, Liu Z, Wang M, Qi L, Jin W, Luo Z, Zhou P, Lee Y, Staskawicz B, He SY, Sun W. Science 2025; 390: 1299-1304. (doi: 10.1126/science.ady8325)
(Science Commentary: Tricks for treats)
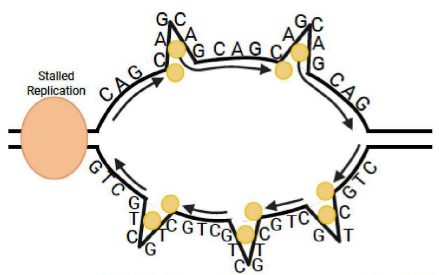
124. REV1 inhibition enhances trinucleotide repeat mutagenesis. Siegel A, Almstead D, Kothandaraman N, Reich J, Lamkin E, Victor JA, Grover A, Ikeh K, Koval H, Crompton A, Jang H, Lee H, Del Rio Guerra R, Korzhnev DM, Kyle Hadden M, Hong J, Zhou P, Chatterjee N. Open Biology 2025; 15: 250234. (doi.org/10.1098/rsob.250234)

123. Mechanism of controlled radical initiation in radical SAM GTP 3′,8-cyclase. Pang H†, Li D†, Wu Q, Zhang P, Yang W, Silakov A*, Zhou P*, Yokoyama K*. PNAS 2025; 122: e2502098122. (doi: 10.1073/pnas.2502098122) †Equal contribution; *Co-correspondence

122. From Obscurity to Opportunity: LpxH Emerges as a Promising Antibiotic Target in the Battle Against Gram-Negative Pathogens. Dome PA†, Cochrane CS†, Switzera HJ†, Lee H, Jeonga P, Hong J*, and Zhou P*. ACS Infectious Diseases 2025; 11: 2993-3008. (doi/10.1021/acsinfecdis.5c00625) †Equal contribution; *Co-correspondence

121. Structural Insights into Proteolysis-Dependent and -Independent Suppression of the Master Regulator DELLA by the Gibberellin Receptor. Dahal P†, Wang Y†, Hu J, Park J, Forker K, Zhang Z, Sharma K, Borgnia MJ, Sun T-p*, Zhou P*. PNAS 2025; 122: e2511012122 (doi: 10.1073/pnas.2511012122) †Equal contribution; *Co-correspondence
Commentary: The molecular structure of the master growth regulator in plants emerges into the spotlight. Yoshida H and Matsuoka M. PNAS 2025; 122(36): e2519807122. (doi: 10.1073/pnas.2519807122)

120. Pathological modulation of genome maintenance by cancer/testes antigens (CTAs). Vaziri C, Forker K, Zhang X, Wu D, Zhou P, Bowser JL DNA Repair (Amst) 2025; 147: 103818. (10.1016/j.dnarep.2025.103818)

119. Assessing the threat of Yersinia pestis harboring a multi-resistant IncC plasmid and the efficacy of an antibiotic targeting LpxC. Lemaitre N, Dewitte A, Rakotomanimana F, Gooden D, Toone E, Rajerison M, Zhou P, Sebbane F. Antimicrob Agents Chemother 2025; 69: e0149724. (doi.org/10.1128/aac.01497-24)
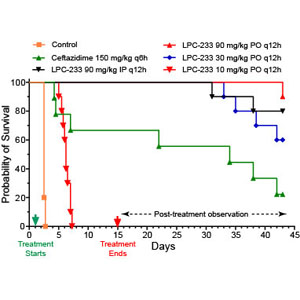
118. Evaluation of a Potent LpxC Inhibitor for Post Exposure Prophylaxis Treatment of Antibiotic Resistant Burkholderia pseudomallei in a Murine Infection Model. Heine HS*, Purcell BK, Duncan C, Miller L, Craig JE, Chase A, Honour L, Vicchiarelli M, Drusano GL, and Zhou P*. Antimicrob Agents Chemother 2025; 69: e0129524 (doi: 10.1128/aac.01295-24) *Co-correspondence

117. Design and Evaluation of Pyridinyl Sulfonyl Piperazine LpxH Inhibitors with Potent Antibiotic Activity Against Enterobacterales. Ennis AF†, Cochrane CS†, Dome PA†, Jeong P, Yu J, Lee H, Williams CS, Ha Y, Yang W, Zhou P* and Hong J*. JACS Au 2024 Nov 11;4(11):4383-4393. (doi: 10.1021/jacsau.4c00731) †Equal Contributions; *Co-correspondence

116. PCNA-binding activity separates RNF168 functions in DNA replication and DNA double-stranded break signaling. Yang Y, Jayaprakash D, Jhujh SS, Reynolds JJ, Chen S, Gao Y, Anand JR, Mutter-Rottmayer E, Ariel P, An J, Cheng X, Pearce KH, Blanchet SA, Nandakumar N, Zhou P, Fradet-Turcotte A, Stewart GS, Vaziri C. Nucleic Acids Res. 2024 Oct 24:gkae918. (doi: 10.1093/nar/gkae918) PMID: 39445802

115. Protocol for Designing De Novo Noncanonical Peptide Binders in OSPREY. Childs H, Guerin N, Zhou P, and Donald BR. J Compt Biol. 2024 Oct; 31(10): 965-974. (link) doi: 10.1089/cmb.2024.0669. PMID: 39364612
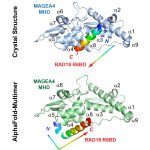
114. Crystal structure of MAGEA4 MHD-RAD18 R6BD reveals a flipped binding mode compared to AlphaFold2 prediction. Forker K, Fleming MC, Pearce KH, Vaziri C, Bowers AA, Zhou P. EMBO J. 2024; 43: 2835-2839. (link) doi: 10.1038/s44318-024-00140-2. PMID: 38907034

113. DexDesign: an OSPREY-based algorithm for designing de novo D-peptide inhibitors. Guerin N, Childs H, Zhou P, Donald BR. Protein Eng Des Sel. 2024; 37: gzae007. (link) https://doi.org/10.1093/protein/gzae007

112. AI is a viable alternative to high throughput screening: a 318-target study. The Atomwise AIMS Program. Scientific Reports Volume 14, Article Number 7526. 2024. (link) https://doi.org/10.1038/s41598-024-54655-z

111. The landscape of small-molecule prodrugs. Fralish, Z., Chen, A., S. et al. Nat Rev Drug Discov. 2024; 23: 365-380. (link) https://doi.org/10.1038/s41573-024-00914-7

110. Bacterial pathogens deliver water- and solute-permeable channels to plant cells. Nomura K†, Andreazza F†, Cheng J†, Dong K*, Zhou P* & He SY*. Nature. 2023; 621: 586-591. (link) https://doi.org/10.1038/s41586-023-06531-5
See news at Eurekalert (†Equal contribution; *Co-correspondence).

109. Preclinical safety and efficacy characterization of an LpxC inhibitor against Gram-negative pathogens. Zhao J†, Cochrane CS†, Najeeb J, Gooden G, Sciandra C, Fan P, Lemaitre N, Newns K, Nicholas RA, Guan Z, Thaden JT, Fowler VG, Spasojevic I, Sebbane F, Toone EJ, Duncan C, Gammans R, and Zhou P*. Science Translational Medicine. 2023; 15: eadf56. (link) https://doi.org/10.1126/scitranslmed.adf5668 (†Equal contribution; *Correspondence)

108. Discovery, characterization, and redesign of potent antimicrobial thanatin orthologs from Chinavia ubica and Murgantia histrionica targeting E. coli LptA. Huynh K#, Kibrom A#, Donald B.R.*, Zhou P*. J Struct Biol X: X. 2023; 8: 100091. (link) https://doi.org/10.1016/j.yjsbx.2023.100091. (#equal contribution; *correspondence)

107. Development of LpxH inhibitors chelating the active site di-manganese metal cluster of LpxH. Kwak SH, Cochrane CS, Cho J, Dome PA, Ennis AF, Kim JH, Zhou P, Hong J. ChemMedChem. 2023 Apr 4;e202300023. (link) https://doi.org/10.1002/cmdc.202300023

106. Structure and dynamics of the Arabidopsis O-fucosyltransferase SPINDLY. Kumar S, Wang Y, Zhou Y, Dillard L, Li FW, Sciandra CA, Sui N, Zentella R, Zahn E, Shabanowitz J, Hunt DF, Borgnia MJ, Bartesaghi A, Sun TP, Zhou P. Nat Commun. 2023 Mar 20;14(1):1538 (link) https://doi.org/10.1038/s41467-023-37279-1
105. Seeing is believing: Understanding functions of NPR1 and its paralogs in plant immunity through cellular and structural analyses. Zhou P, Zavaliev R, Xiang Y, Dong X. Curr Opin Plant Biol. 2023 Mar 17;73:102352. (link) https://https://doi.org/10.1016/j.pbi.2023.102352

104. Roles of trans-lesion synthesis (TLS) DNA polymerases in tumorigenesis and cancer therapy. Anand J, Chiou L, Sciandra C, Zhang X, Hong J, Wu D, Zhou P, Vaziri C. NAR Cancer 2023; 5, 1-23. (link) https://doi.org/10.1093/narcan/zcad005

103. From magic spot ppGpp to MESH1: Stringent response from bacteria to metazoa. Chi JT, Zhou P. PLoS Pathog. 2023 Feb 2;19(2):e1011105. doi: 10.1371/journal.ppat.1011105. (link) https://doi.org/10.1371/journal.ppat.1011105
102. Metazoan stringent-like response mediated by MESH1 phenotypic conserva‐ tion via distinct mechanisms. Mestre AA, Zhou P, and Chi JT. Comput Struct Biotechnol J. 2022 May 6;20:2680-2684 (link) https://doi.org/10.1016/j.csbj.2022.05.001

101. Structural basis of NPR1 in activating plant immunity. Kumar S, Zavaliev R, Wu Q, Zhou Y, Cheng J, Dillard L, Powers J, Withers J, Zhao J, Guan Z, Borgnia MJ, Bartesaghi A, Dong X & Zhou P. Nature 2022 May (link) (Also see coverage on the Howard Hughes Medical Institute website) https://doi.org/10.1038/s41586-022-04699-w

100. MESH1 Knockdown Triggers Proliferation Arrest Through TAZ Repression. Sun T, Ding CKC, Zhang Y, Zhang Y, Lin CC, Wu J, Setayeshpour Y, Coggins S, Shepard C, Macias E, Kim B, Zhou P, Gordân R & Chi JT Cell Death & Disease 2022 March (link) doi:10.3389/fmolb.2021.758228

99. Degradation of Components of the Lpt Transenvelope Machinery Reveals LPS-Dependent Lpt Complex Stability in Escherichia coli. Martorana AM, Moura ECCM, Sperandeo P, Vincenzo FD, Liang X, Toone E, Zhou P, Polissi A. Front Mol Biosci. 2021: 8: 758228. (link) doi:10.3389/fmolb.2021.758228

98. Adaptive responses of Pseudomonas aeruginosa to treatment with antibiotics. Wüllner D, Gesper M, Haupt A, Liang X, Zhou P, Dietze P, Narberhaus F, Bandow JE. Antimicrob Agents Chemother. 2021 November (link) doi: 10.1128/AAC.00878-21

97. REV1 Inhibition Enhances Radioresistance and Autophagy. Ikeh KE, Lamkin EN, Crompton A, Deutsch J, Fisher KJ, Gray M, Argyle DJ, Lim WY, Korzhnev DM, Hadden MK, Hong J,Zhou P and Chatterjee N. Cancers 2021 September (link) doi:10.1038/s41467-021-25909-5

96. Direct Photoresponsive Inhibition of a p53-Like Transcription Activation Domain in PIF3 by Arabidopsis Phytochrome B . Yoo CY, He J, Sang Q, Qiu Y, Long L, Kim RJ, Chong EG, Hahm J, Morffy N, Zhou P, Strader LC, Nagatani A, Mo B, Chen X, Chen M. Nat Commun. 2021 September (link) doi:10.1038/s41467-021-25909-5
95. Mutations in PBP2 From Ceftriaxone-Resistant Neisseria Gonorrhoeae Alter the Dynamics of the b3-b4 Loop to Favor a Low-Affinity Drug-Binding State. Fenton BA, Tomberg J, Sciandra CA, Nicholas RA, Davies C, Zhou P J Biol Chem. 2021 September (link) doi: 10.1038/s41467-021-25909-5

94. SARS-CoV-2 triggers DNA damage response in Vero E6 cells. Victor J, Deutsch J, Whitaker A; Lamkin E; March A; Zhou P, Botten JW, Chatterjee N. Biochem. Biophys. Res. Commun. 2021 September (link) doi:10.1101/2021.09.08.459535
93. Oncogenic KRAS is dependent upon an EFR3A-PI4KA signaling axis for potent tumorigenic activity. Adhikari H, Kattan WE, Kumar S, Zhou P, Hancock JF, Counter CM. Nat Commun. 2021 September (link) doi:10.1038/s41467-021-25523-5

92. The Regulation of Ferroptosis by MESH1 Through the Activation of the Integrative Stress Response. Lin CC, Ding CC, Sun T, Wu J, Chen KY, Zhou P, Chi JT. Cell Death Dis. 2021 July (link) doi:10.1038/s41419-021-04018-7.
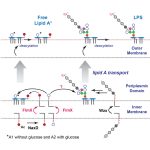
91. Francisella Flmx Broadly Affects Lipopolysaccharide Modification and Virulence. Chin CY, Zhao J, Llewellyn AC, Golovliov I, Sjöstedt A, Zhou P, Weiss DS. Cell Rep. 2021 June (link) doi:10.1016/j.celrep.2021.109247.

90. Structure- and Ligand-Dynamics-Based Design of Novel Antibiotics Targeting Lipid A Enzymes LpxC and LpxH in Gram-Negative Bacteria. Zhou P, Hong J. Acc Chem Res. 2021 March (link) doi: 10.1021/acs.accounts.0c00880.

89. REV1 inhibitor JH-RE-06 enhances tumor cell response to chemotherapy by triggering senescence hallmarks. Chatterjee N, Whitman MA, Harris CA, Min SM, Jonas O, Lien EC, Luengo A, Vander Heiden MG, Hong J, Zhou P, Hemann MT, Walker GC. PNAS 2020. (link) doi:10.1073/pnas.2016064117.

88. Synthesis and evaluation of sulfonyl piperazine LpxH inhibitors. Kwak SH, Cochrane CS, Ennis AF, Lim WY, Webster CG, Cho J, Fenton BA, Zhou P, Hong J. Bioorg Chem. 2020 Jun 30;102:104055.
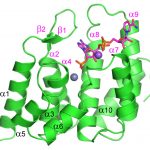
87. MESH1 is a cytosolic NADPH phosphatase that regulates ferroptosis. Ding CKC, Rose J, Sun T, Wu J, Chen PH, Lin CC, Yang WH, Chen KY, Lee H, Xu E, Tian S, Akinwuntan K, Zhao J, Guan Z, Zhou P & Chi JT. Nature Metabolism 2020; 2: 270-277 (link) doi.org/10.1038/s42255-020-0181-1

86. Structural basis of the UDP-diacylglucosamine pyrophosphohydrolase LpxH inhibition by sulfonyl piperazine antibiotics. Cho J#, Lee M#, Cochrane CS#, Webster CG, Fenton BA, Zhao J, Hong J*, and Zhou P*. PNAS 2020; 117: 4109-4116. (link)

85. Metabolic engineering of Escherichia coli to produce a monophosphoryl lipid A adjuvant. Ji Y, An J, Hwang D, Ha DH, Lim SM, Lee C, Zhao J, Yang EG, Song HK, Zhou P, Cheng HS. Metabolic Engineering, 2020; 57: 193-202. (PDF)

84. The Lipid A 1-Phosphatase, LpxE, Functionally Connects Multiple Layers of Bacterial Envelope Biogenesis. Zhao J#, An J#, Hwang D#, Wu Q#, Wang S, Gillespie RA, Yang EG, Guan Z, Zhou P*, Chung HS*. mBio 2019; 10: e00886-19. (PDF)

83. NCP activates chloroplast transcription bycontrolling phytochrome-dependent dual nuclearand plastidial switches. Yang EJ#, Yoo CY#, Liu J#, Wang H, Cao J, Li F-W, Pryer KM, Sun T-P, Weigel D, Zhou P*, Chen M*. Nature Communications 2019; 10: 2630.(PDF) [Also read, "It is not easy being green" -- the story of NCP (NUCLEAR CONTROL OF PEP ACTIVITY) and its cousin RCB (REGULATOR OF CHLOROPLAST BIOGENESIS), also published in Nature Communications]

82. A Small Molecule Targeting Mutagenic Translesion Synthesis Improves Chemotherapy. Wojtaszek JL#, Chatterjee N#, Najeeb J#, Ramos A, Lee M, Bian K, Xue JY, Fenton BA, Park H, Li D, Hemann MT*, Hong J*, Walker GC*, Zhou P*. Cell (2019). (link) (Also see coverage on Science Daily and EurekAlert!)

81. Structure−Activity Relationship of Sulfonyl Piperazine LpxH Inhibitors Analyzed by an LpxE-Coupled Malachite Green Assay. Lee M, Zhao J, Kwak S-H, Cho J, Lee M, Gellespie TA, Kwon D-Y, Lee H, Wu Q, Zhou P*, and Hong J*. ACS Infectious Diseases 2019; 5: 641-651 (PDF).
79. Lipid A Has Significance for Optimal Growth of Coxiella burnetii in Macrophage-Like THP-1 Cells and to a Lesser Extent in Axenic Media and Non-phagocytic Cells. Wang T, Yu Y, Liang X, Luo S, He Z, Sun Z, Jiang Y, Omsland A, Zhou P, Song L. Front Cell Infect Microbiol. 2018 Jun 8;8:192. doi: 10.3389/fcimb.2018.00192. eCollection 2018.

78. A pH-gated conformational switch regulates the phosphatase activity of bifunctional HisKA-family histidine kinases. Liu Y, Rose J, Huang S, Hu Y, Wu Q, Wang D, Li C, Liu M, Zhou P, Jiang L. Nat Commun. 2017; 8(1). 2104. (doi: 10.1038/s41467-017-02310-9)

77. Curative Treatment of Severe Gram-Negative Bacterial Infections by a New Class of Antibiotics Targeting LpxC. Lemaître N, Liang X, Najeeb J, Lee CJ, Titecat M, Leteurtre E, Simonet M, Toone EJ, Zhou P, Sebbane F. MBio. 2017 Jul 25;8(4). pii: e00674-17. (PDF)

76. Probing the excited-state chemical shifts and exchange parameters by nitrogen-decoupled amide proton chemical exchange saturation transfer (HNdec-CEST). Wu Q, Fenton B, Wojtaszek J, and Zhou P. Chemical Communications, 2017, DOI: 10.1039/C7CC05021F. (PDF)

75. (Book Chapter of Fast NMR Data Acquisition). Chapter 6: Backprojection and Related Methods. Brian E. Coggins and Pei Zhou Chapter 7: CLEAN. Brian E. Coggins and Pei Zhou

74. The Arabidopsis O-fucosyltransferase SPINDLY activates nuclear growth repressor DELLA. Zentella R, Sui N, Barnhill B, Hsieh WP, Hu J, Shabanowitz J, Boyce M, Olszewski NE, Zhou P, Hunt DF, Sun TP. Nat Chem Biol. 2017 Feb 28. doi: 10.1038/nchembio.2320. PMID: 28244988 (PDF)

73. Structure, inhibition, and regulation of essential lipid A enzymes. Zhou P and Zhao J. Biochim Biophys Acta. 2016 Dec 8. pii: S1388-1981(16)30331-6. doi: 10.1016/j.bbalip.2016.11.014. Review. PMID: 27940308

72. Structure of the essential Haemophilus influenzae UDP-diacylglucosamine pyrophosphohydrolase LpxH in lipid A biosynthesis. Cho J, Lee C-J, Zhao J, Young HE, Zhou P. Nature Microbiology 1, Article number: 16154 (2016) doi:10.1038/nmicrobiol.2016.15472.

71. Unbiased measurements of reconstruction fidelity of sparsely sampled magnetic resonance spectra. Wu Q, Coggins BE, Zhou P. Nature Communications 2016; 7: 12281 (link)
 70. High susceptibility of multidrug- and extensively drug-resistant Gram-negative pathogens to biphenyl-diacetylene-based difluoromethyl-allo-threonyl-hydroxamate LpxC inhibitors. Titecat M, Liang X, Lee C-J, Charlet A, Hocquet D, Lambert T, Jean-Marie P, Courcol RJ, Sebbane F, Toone E, Zhou P, Lemaitre N. Journal of Antimicrobial Chemotherapy 2016 Jun 20. pii: dkw210. (link)
70. High susceptibility of multidrug- and extensively drug-resistant Gram-negative pathogens to biphenyl-diacetylene-based difluoromethyl-allo-threonyl-hydroxamate LpxC inhibitors. Titecat M, Liang X, Lee C-J, Charlet A, Hocquet D, Lambert T, Jean-Marie P, Courcol RJ, Sebbane F, Toone E, Zhou P, Lemaitre N. Journal of Antimicrobial Chemotherapy 2016 Jun 20. pii: dkw210. (link)
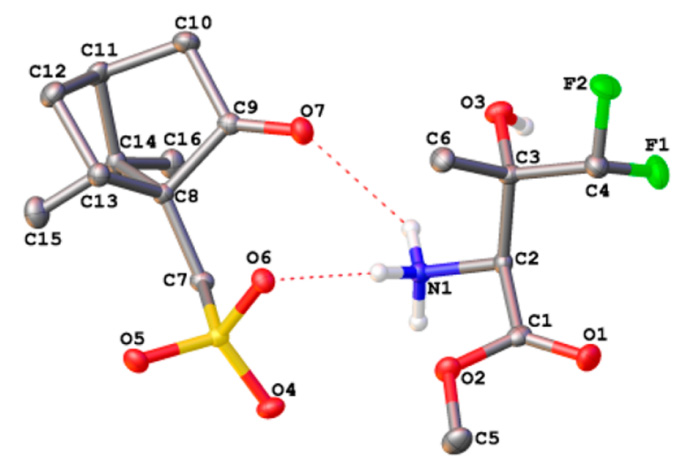
69. A Scalable Synthesis of the Difluoromethyl-allo-threonyl hydroxamate-based LpxC Inhibitor LPC-058. Liang X, Gopalaswamy R, Navas Iii F, Toone EJ, Zhou P. J Org Chem. 2016 Apr 29. doi:10.1021/acs.joc.6b00589.

68. Discovery of the elusive UDP-diacylglucosamine hydrolase in the lipid A biosynthetic pathway in Chlamydia trachomatis. Young HE, Zhao J, Barker JR, Guan Z, Valdivia RH, Zhou P. MBio. 2016; 7(2). pii: e00090-16. doi: 10.1128/mBio.00090-16.
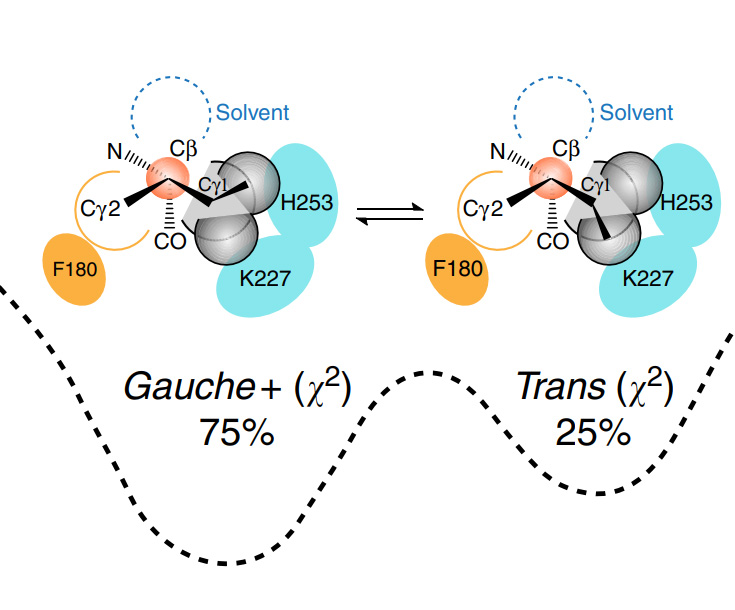
67. Drug design from the cryptic inhibitor envelope. Lee C-J, Liang X, Wu Q, Najeeb J, Zhao J, Gopalaswamy R, Titecat M, Sebbane F, Lemaitre N, Toone EJ, Zhou P. Nature Communications 2016; 7: 10638. (link)

65. Systematic solution to homo-oligomeric structures determined by NMR. Martin J, Zhou P, Donald BR. PROTEINS: Structure, Function, and Bioinformatics 2015; 83: 651-61(PDF)
63. Structural basis of lipid binding for the membrane-embedded tetraacyldisaccharide-1-phosphate 4'-kinase LpxK. Emptage RP, Tonthat NK, York JD, Schumacher MA, and Zhou P. J. Biol. Chem. 2014 Aug 29;289(35):24059-68. doi:10.1074/jbc.M114.589986. (PDF)
62. A CRISPR-Cas system enhances envelope integrity mediating antibiotic resistance and inflammasome evasion. Sampson TR, Napier BA, Schroeder MR, Louwen R, Zhao J, Chin C-Y, Ratner HK, Llewellyn AC, Jones CL, Laroui H, Merlin D, Zhou P, Endtz HP, and Weiss DS. PNAS 2014; 11163-11168. doi:10.1073/pnas.1323025111. (PDF)
61. Sparsely-sampled, high-resolution 4-D omit spectra for detection and assignment of intermolecular NOEs of protein complexes. Wang S and Zhou P. J Biomol NMR 2014; 59: 51-6. (PDF)
60. Chasing acyl carrier protein through a catalytic cycle of lipid A production. Masoudi A, Raetz CRH, Zhou P, and Pemble CW. Nature. 2014 Jan 16; 505(7483): 422-6. (PDF)
59. Structural Basis of the Promiscuous Inhibitor Susceptibility of E. coli LpxC. Lee C-J, Liang X, Gopalaswamy R, Najeeb J, Ark, ED, Toone EJ, and Zhou P. ACS Chem. Biol. 2014 Jan 17;9(1):237-46. doi: 10.1021/cb400067g. (PDF)
57. Synthesis, structure and antibiotic activity of aryl-substituted LpxC inhibitors. Liang X, Lee C-J, Zhao J, Toone EJ, and Zhou P. J. Med. Chem., 2013; 56(17): 6954-66, (PDF)
56. The UDP-diacylglucosamine pyrophosphohydrolase LpxH in lipid A biosynthesis utilizes A Mn2+ cluster for catalysis. Young HE, Donohue MP, Smirnova TI, Smirnov AI, and Zhou P. J. Biol. Chem. 2013; 288(38): 26987-7001 (PDF)
55. Storage of hydrogen spin polarization in long-lived (13)c2 singlet order and implications for hyperpolarized magnetic resonance imaging. Feng Y, Theis T, Liang X, Wang Q, Zhou P, Warren WS. JACS. 2013; 135(26): 9632-5. (link)
54. Mechanistic characterization of the tetraacyldisaccharide-1-phosphate 4´-kinase LpxK involved in lipid A biosynthesis. Emptage RP, Pemble CW, York JD, Raetz CR, Zhou P. Biochemistry. 2013; 52, 2280-90. (PDF)
53. Specific interaction of the transcription elongation regulator TCERG1 with RNA polymerase II requires simultaneous phosphorylaHASH: a Program to Accurately Predict Protein Ha Shifts from Neighboring Backbone tion at Ser2, Ser5 and Ser7 within the carboxyl-terminal domain repeat. Liu J, Fan S, Lee CJ, Greenleaf AL, Zhou P. J Biol Chem. 2013; 288, 10890-901. (PDF)
52. Mutants resistant to LpxC inhibitors by rebalancing cellular homeostasis. Zeng D, Zhao J, Chung HS, Guan Z, Raetz CR, Zhou P. J Biol Chem. 2013; 288: 5475-86. (PDF)
51. HASH: a Program to Accurately Predict Protein Ha Shifts from Neighboring Backbone Shifts. Zeng J, Zhou P, and Donald BR. J Biomol NMR 2013; 55: 105-18. (PDF)
49. Structural basis of Rev1-mediated assembly of a quaternary vertebrate translesion polymerase complex consisting of Rev1, heterodimeric Pol zeta and Pol kappa. Wojtaszek J, Lee CJ, D'Souza S, Minesinger B, Kim H, D'Andrea AD, Walker GC, and Zhou P. J Biol Chem. 2012; 287(40): 33836-46 (PDF) (Recommended by Faculty of 1000)
48. Multifaceted recognition of vertebrate Rev1 by translesion polymerases zeta and kappa. Wojtaszek J, Liu J, D'Souza S, Wang S, Xue Y, Walker GC, Zhou P. J Biol Chem. 2012 July 27; 287 (31): 26400-8. (PDF)
47. Efficient acquisition of high-resolution 4-D diagonal-suppressed methyl-methyl NOESY for large proteins. Wen J, Zhou P, Wu J. J Magn Reson. 2012 May;218:128-32. PMID: 22464875. (PDF)

46. Protein loop closure using orientational restraints from NMR data.Tripathy C, Zeng J, Zhou P, Donald BR. Proteins. 2011 Sep 26. doi: 10.1002/prot.23207. (PDF)
45. A geometric arrangement algorithm for structure determination of symmetric protein homo-oligomers from NOEs and RDCs. Martin JW, Yan AK, Bailey-Kellogg C, Zhou P, Donald BR. J Comput Biol. 2011 Nov;18(11):1507-23. Epub 2011 Oct 28.
44. A Bayesian approach for determining protein side-chain rotamer conformations using unassigned NOE data. Zeng J, Roberts KE, Zhou P, Donald BR. J Comput Biol. 2011 Nov;18(11):1661-79.
43. 1H, 13C and 15N backbone and side-chain resonance assignments of Drosophila melanogaster Ssu72. Werner-Allen JW, Zhou P. Biomol NMR Assign. 2011 Jul 6. (PDF)
42. Protein Side-Chain Resonance Assignment and NOE Assignment Using RDC-Defined Backbones without TOCSY Data. Zeng J, Zhou P and Donald BR. J Biomol NMR. 2011 Jun 25. (PDF)
41. Lipooligosaccharide is required for the generation of infectious elementary bodies in Chlamydia trachomatis. Nguyen BD, Cunningham D, Liang X, Chen X, Toone EJ, Raetz CR, Zhou P, Valdivia RH. Proc Natl Acad Sci U S A. 2011 Jun 21;108(25):10284-9. (PDF)
40. A graphical method for analyzing distance restraints using residual dipolar couplings for structure determination of symmetric protein homo-oligomers. Martin JW, Yam AK, Bailey-Kellogg C, Zhou P and Donald BR. Protein Sci 2011; 20: 970-85. (PDF)

39. Sparsely-sampled high-resolution 4-D experiments for efficient backbone resonance assignment of disordered proteins. Wen J, Wu J, and Zhou P. J Magn Reson 209, 94-100 (2011). (PDF)

38. Cis proline-mediated pSer5-dephosphorylation by the RNA polymerase II CTD phosphatase Ssu72. Werner-Allen JW, Lee CJ, Liu P, Nicely NI, Wang S, Greenleaf AL, Zhou P. Journal of Biological Chemistry 286, 5717-5826 (2011). (PDB: 3P3Y) (PDF)

37. Syntheses, structures and antibiotic acitivities of LpxC inhibitors based on the diacetylene scaffold. Liang X, Lee CJ, Chen X, Chung HS, Zeng D, Raetz CRH, Li Y, Zhou P, Toone EJ. Bioorganic & Medicinal Chemistry 19: 852-60 (2011). (PDB: 3PS1, 3PS2, 3PS3) (PDF)

36. Species-specific and inhibitor-dependent conformations of LpxC-Implications for antibiotic design. Lee CJ, Liang X, Chen X, Zeng D, Joo SH, Chung HS, Barb AW, Swanson SM, Nicholas RA, Li Y, Toone EJ, Raetz CRH, Zhou P. Chemistry and Biology 18: 38-47 (2011). (PDB: 3P3C, 3P3E, 3P3G) (PDF)

35. HDAC6 and Ubp-M BUZ Domains Recognize Specific C-Terminal Sequences of Proteins. Hard RL, Liu J, Shen J, Zhou P, Pei D. Biochemistry 49:10737-46 (2010). (PDF)

34. The unusual UBZ domain of Saccharomyces cerevisiae polymerase eta. Woodruff RV, Bomar MG, D'Souza S, Zhou P, Walker GC. DNA Repair 9: 1130-41 (2010). (PDF)

33. Radial Sampling for Fast NMR: Concepts and Practices Over Three Decades. Coggins BE, Venters RA, and Zhou P. Progress in Nuclear Magnetic Resonance Spectroscopy 57: 381-419 (2010). (Review)

32. Fast Acquisition of High Resolution 4-D Amide-Amide NOESY with Diagonal Suppression, Sparse Sampling and FFT-CLEAN. Werner-Allen JW, Coggin BE and Zhou P. J Magn Reson 204,:173-178. (2010) (PDF)

31. Unconventional Ubiquitin Recognition by the Ubiquitin-Binding Motif within the Y Family DNA Polymerases i and Rev1. Bomar MG, D’Souza S, Bienko M, Dikic I, Walker GC, and Zhou P. Molecular Cell 37, 408-417. (2010) (PDF)
30. Assignment of 1H, 13C and 15N backbone resonances of Escherichia coli LpxC bound to L-161,240. Barb AW, Jiang L, Raetz CR and Zhou P. Biomol NMR Assign. (PMID: 19941092) (PDF)
29. Overcoming the Solubility Limit with Solubility-Enhancement Tags: Successful Applications in Biomolecular NMR Studies. Zhou P and Wagner G. J Biomolecular NMR 46: 23-31. (2010) (Review)
28. High-resolution protein structure determination starting with a global fold calculated from exact solutions to the RDC equations. Zeng J, Boyles J, Tripathy C, Wang L, Yan A, Zhou P and Donald BR. J Biomolecular NMR45: 265-81. (2009) (PDF)
27. Uridine-based inhibitors as new leads for antibiotics targeting E. coli LpxC. Barb AW, Leavy TM , Robins L, Guan Z , Six D, Zhou P, Bertozzi C and Raetz CRH. Biochemistry 48, 3068-77 (2009). (PDF)
25. Mechanism and inhibition of LpxC: an essential zinc-dependent deacetylase of bacterial lipid A synthesis. Barb AW and Zhou P. Curr Pharm Biotechnol 9, 9-15 (2008). (Review)
24. High resolution 4-D spectroscopy with sparse concentric shell sampling and FFT-CLEAN. Coggins BE and Zhou P.J Biomol NMR 42, 225–239 (2008). (PDF)
23. Structure of the deacetylase LpxC bound to the antibiotic CHIR-090: time-dependent inhibition and specificity in ligand binding. Barb A, Jiang L, Raetz CRH and Zhou P.Proc Natl Acad Sci USA 104, 18433-18438 (2007). (PDF)
22. The ADAM10 prodomain is a specific inhibitor of ADAM10 proteolytic activity and inhibits cellular shedding events. Moss ML, Bomar M, Liu Q, Sage H, Dempsey P, Lenhart PM, Gillispie PA, Stoeck A, Wildeboer D, Bartsch JW, Palmisano R, Zhou P.J Biol Chem282, 35712-35721 (2007). (PDF)
21. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SCR and Lin H.Genes & Development21, 2300-2311 (2007).
20. The low affinity IgE receptor (CD23) is cleaved by the metalloproteinase ADAM10. Lemieux GA, Blumenkron F, Yeung N, Zhou P, Williams J, Grammer AC, Petrovich R, Lipsky PE, Moss ML, Werb Z. J Biol Chem282, 14836-44 (2007). (PDF)
19. Solution Structure of the Ubp-M BUZ Domain, a Highly Specific Protein Module that Recognizes the C-terminal Tail of Free Ubiquitin. Pai MT, Tzeng SR, Kovacs JJ, Keaton MA, Li SS, Yao TP, Zhou P. J Mol Biol370,290-302 (2007). (PDF)
18. Inhibition of Lipid A Biosynthesis as the Primary Mechanism of CHIR-090 Antibiotic Activity inEscherichia coli. Barb AW, McClerren AL, Snehelatha K, Reynolds CM, Zhou P, Raetz CRH. Biochemistry46, 3793-3802 (2007). (PDF)
17. Structure of the ubiquitin-binding zinc finger domain of human DNA Y-polymeraseh. Bomar MG, Pai M, Tzeng S, Li S and Zhou P.EMBO reports 8, 247-251 (2007). (PDF) (Supplementary Information)
16. Sampling of the NMR time domain along concentric rings. Coggins BE and Zhou P.J Magn Reson, 184, 207-221 (2007). (PDF)
15. Fourier Transforms of Radially-Sampled NMR Data. Coggins BE and Zhou P.J Magn Reson182, 84-95 (2006). (PDF)
14. A 'just-in-time' HN(CA)CO experiment for the backbone assignment of large proteins with high sensitivity. Werner-Allen JW, Jiang L, and Zhou P.J Magn Reson181, 177-180 (2006). (PDF)
13. PR-CALC: A Program for the Reconstruction of NMR Spectra from Projections. Coggins BE and Zhou P.J Biomol NMR 34, 179-95 (2006). (PDF) (Supplementary Material)
12. Evaluating the quality of NMR structures by local density of protons. Ban YA, Rudolph J, Zhou P and Edelsbrunner H.Proteins: Structure, Function, and Bioinformatics62, 852-864 (2006). (PDF)
10. Filtered Backprojection for the Reconstruction of a High-Resolution (4,2)D CH3-NH NOESY Spectrum on a 29 kDa Protein. Coggins BE, Venters RA and Zhou P.J Am Chem Soc 127, 11562-11563 (2005). (PDF) (Supplementary Materials)
9. (4,2)D Projection-Reconstruction Experiments for Protein Backbone Assignment: Application to Human Carbonic Anhydrase II and Calbindin D28K. Venters RA, Coggins BE, Kojetin D, Cavanagh J and Zhou P. J Am Chem Soc 127, 8785-8795 (2005). (PDF) (Supplementary Material)
8. Rapid assignment of protein side chain resonances using projection-reconstruction of (4,3)D HC(CCO)NH and intra-HC(C)NH experiments. Jiang L, Coggins BE and Zhou P.J Magn Reson 175, 170-176 (2005). (PDF)
7. Refined Solution Structure of the LpxC-TU-514 Complex and pKa Analysis of an Active Site Histidine: Insights into the Mechanism and Inhibitor Design. Coggins BE, McClerren AL, Jiang L, Li X, Rudolph J, Hindsgaul O, Raetz CRH, and Zhou P.Biochemistry 44, 1114-1126 (2005). (PDF)
6. Kinetic Analysis of the Zinc-Dependent Deacetylase in the Lipid A Biosynthetic Pathway. McClerren AL, Zhou P, Guan Z, Raetz CRH, and Rudolph J.Biochemistry 44, 1106-1113 (2005). (PDF)
5. Assignment of the 1H, 13C and 15N Resonances of the LpxC Deacetylase from Aquifex aeolicus in Complex with the Substrate-Analog Inhibitor TU-514. Coggins BE, Li X, Hindsgaul O, Raetz CRH, Zhou P. J Biomol NMR 28, 201-202 (2004). PubMed Abstract
4. Generalized Reconstruction of n-D NMR Spectra from Multiple Projections: Application to the 5-D HACACONH Spectrum of Protein G B1 Domain. Coggins BE, Venters RA, Zhou P.J Am Chem Soc 126, 1000-1001 (2004). (PDF)
3. Structure of the LpxC deacetylase with a bound substrate-analog inhibitor. Coggins BE, Li X, McClerren AL, Hindsgaul O, Raetz CRH, Zhou P.Nat Struct Biol 10, 645-651 (2003). (PDF)
2. Characteristics of the Interaction of a Synthetic Human Tristetraprolin Tandem Zinc Finger Peptide with AU-rich Element-containing RNA Substrates. Blackshear PJ, Lai WS, Kennington EA, Brewer G, Wilson GM, Guan X and Zhou P.J Biol Chem 278,19947-19955 (2003). (PDF)
1. PACES: Protein sequential assignment by computer-assisted exhaustive search. Coggins BE and ZhouP. J Biomol NMR 26, 93-111 (2003). (PDF)
P7. A novel approach for characterizing protein ligand complexes: molecular basis for specificity of small-molecule Bcl-2 inhibitors. Lugovskoy AA, Degterev AI, Fahmy AF, Zhou P, Gross JD, Yuan J, Wagner G. J Am Chem Soc. 2002 Feb 20;124(7):1234-40. doi: 10.1021/ja011239y. PMID: 11841292
P6. A solubility-enhancement tag (SET) for NMR studies of poorly behaving proteins. Zhou P, Lugovskoy AA and Wagner G. J Biomolecular NMR 20, 11-14 (2001). (PDF)
P5. Solution structure of DFF40 and DFF45 N-terminal domain complex and mutual chaperone activity of DFF40 and DFF45. Zhou P, Lugovskoy AA, McCarty JS, Li P and Wagner G. Proc. Natl. Acad. Sci. USA 98, 6051-6055 (2001). (PDF)
P4. Solution structure of the CIDE-N domain of CIDE-B and a model for CIDE-N/CIDE-N interactions in the DNA fragmentation pathway of apoptosis. Lugovskoy AA, Zhou P, Chou JJ, McCarty JS, Li P and Wagner G. Cell 99, 747-755 (1999.) (PDF)
P3. Solution structure of Apaf-1 CARD and its interaction with caspase-9 CARD: A structural basis for specific adaptor/caspase interaction. Zhou P, Chou JJ, Olea RS, Yuan J and Wagner G. Proc. Natl. Acad. Sci. USA 96, 11265-11270 (1999). (PDF)
P2. Solution Structure of the Core NFATC1/DNA complex. Zhou P, Sun LJ, Dötsch V, Wagner G and Verdine GL. Cell 92, 687-696 (1998). (PDF)
P1. Unusual Rel-like Architecture in the DNA-binding Domain of the Transcription Factor NFATc. Wolfe SA, Zhou P, Dötsch V, Chen L, You A, Ho S, Crabtree GR, Wagner G, and Verdine GL. Nature 385, 172-176 (1997). (PDF)

