The Functional Genomics Shared Resource provides reagents, expertise and infrastructure for the application of functional genomic technologies to support the research programs of investigators at Duke and beyond. Through investments in genetic and chemical perturbation technologies, we have assembled state-of-the-art tools, enabling high-throughput functional studies using CRISPR, RNAi and compound libraries. We also provide targeted CRISPR services for custom knockout and knockin cell lines.
- Expertise: The facility provides expertise in the development and use of reagents for genetic perturbation, including CRISPR, RNAi and ORF overexpression.
- Genetic Reagents: We generate and validate custom sgRNA vectors for CRISPR editing of individual genes. We provide assistance with creating CRISPR KO lines as polyclonal pools or clonal KO lines. We also distribute individual lentiviral shRNA clones from the Sigma Mission collection as bacterial glycerol stocks, through our shRNA request system.
- Chemical Reagents: We have chemical collections of varying sizes and purposes available for screening, see https://sites.duke.edu/functionalgenomics/libraries/compound-libraries/ for more details.
- Genome-wide screens: The Functional Genomics Shared Resource has the infrastructure necessary to conduct high-throughput screens, with automated liquid handlers, cell platers and plate washers. Screens can be performed in plate-based assays or in pooled format using a selection-based strategy with any of our CRISPR or shRNA libraries. We also provide assistance in assembling and screening custom CRISPR libraries.
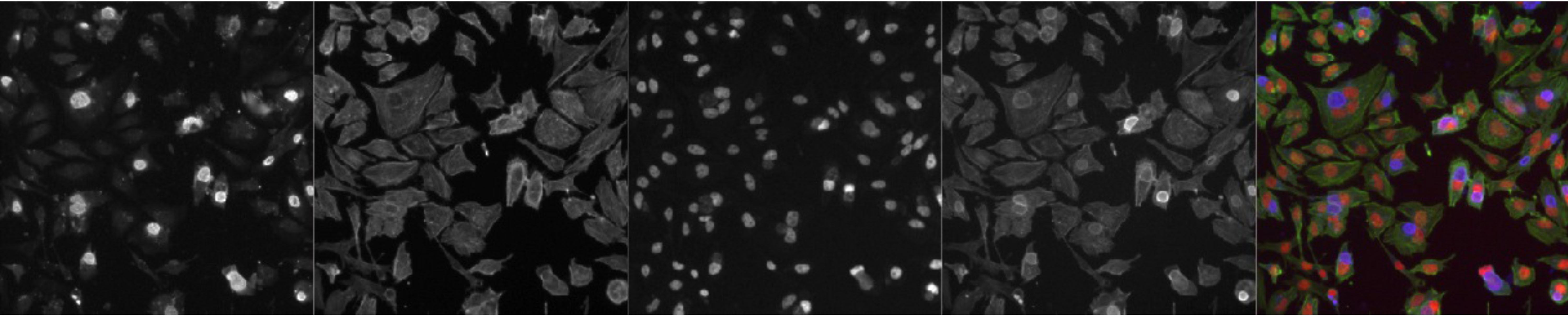
- Phenotypic assays: For plate-based quantitative assays, we have the CellInsight CX5 high-content screening system for image-based analysis and the BMG multimodal plate reader for standard detection assays. All systems are equipped with plate stackers and barcode readers for walk-away functionality and high-throughput processing.