Crystal Structures of Kinesin and Ncd Motor Domains
Structure Determination
Amino acids 1-349 of a human kinesin motor domain were crystallized from bacterially-expressed protein in the presence of MgATP. Crystals were thin rods (1000 x 150 x 40µm) of the space group P212121 with one kinesin and one MgADP per asymmetric unit. Phases were determined using multiple isomorphous replacement (MIR) from two derivatives, 2′-iodo-ATP (I-ATP) and ethylmercurothiosalycilate (EMTS). The atomic model is refined to 1.8Å. The first 6 aa, the last 24 aa, and residues 238-254 are not visible in the refined structure. MgADP is visible in the active site.
Bacterially-expressed Ncd motor domain (residues 335-700) was crystallized in the presence of MgATP. Crystals were of the space group I222 and contain one molecule of MgADP-Ncd in the asymmetric unit. The structure was determined from native Ncd crystals and five isomorphous heavy atom derivatives. The current atomic model of the Ncd motor domain is refined to 2.5Å with an R value of 22.4% and includes 323 amino acid residues, MgADP and 66 water molecules. The first 10 N-terminal and last 33 C-terminal residues were not visible in the electron density maps.
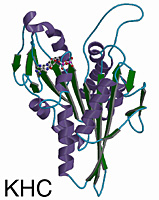
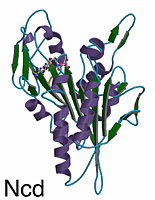
Overview of the Structure
The kinesin and Ncd motor domains resemble asymmetric arrowheads with dimensions of approximately 75 x 45 x 45Å. The bound MgADP lies in an exposed surface cleft. The core of the motor domain is an eight-stranded beta-sheet, flanked by three alpha-helices on each side (see figures). The beta-strand at the right edge of the sheet is unusual in that it is interrupted by an insertion of 26 amino acids. The helix after the P-loop is unusual in that it contains a 9 aa insertion (aa 97-104) flanked by two conserved glycines. This “hairpin” loop extends perpendicular to the helix, but leaves the overall helical path of alpha2 unperturbed.
Comparison of Kinesin and Ncd
Although Ncd and kinesin move in opposite directions along microtubules, their 3-D structures are remarkably similar. The eight-stranded beta-sheet and six alpha-helices of the core, and the three anti-parallel beta-strands that form the small lobe are essentially identical in length, position, and orientation in both motors. Superposition of the core secondary structural elements (146 C-alpha atoms that constitute the central beta-sheet and six helices) gives a root-mean-square (rms) deviation of 1.21Å C-alpha atoms corresponding to 64 central beta-sheet residues can be aligned with a rms deviation of 0.69Å. The position of the MgADP is virtually identical (rms deviation of 0.74Å) in both motors.
The largest differences between kinesin and Ncd are in the surface loops near the nucleotide-binding pocket (L1, L5 and L9), in L2 of the small lobe, and in loop L11 (switch II loop). The N- and C-termini of the Ncd and the kinesin motor domains are expected to show differences, since opposite ends of the polypeptide chains are attached to the coiled coil stalks. Strikingly, the N- and C-termini of the Ncd and kinesin motor domains are positioned similarly in space and located 9Å from one another on the side of the motor opposite to the nucleotide. Electron densities for the first 10 residues of Ncd and 5 residues of kinesin, and last 33 residues of Ncd and 24 residues of kinesin are missing, indicating that these regions are flexible or disordered.
Comparison of Kinesins and Myosins
Secondary structure elements of kinesin and myosin overlap one another in a striking manner. Remarkably, 7 of the 8 core beta-strands (the interrupted edge strand beta5 being the exception) and all six major helices of the kinesin motor domain overlap corresponding structural elements in myosin. The r.m.s deviation of 183 aa alpha-carbon positions in the overlapping regions is 3.5Å. This result was unexpected, given the much larger size of the myosin motor domain and the lack of sequence similarity between the two proteins. The region of myosin that overlaps the kinesin core is confined to the catalytic domain surrounding the nucleotide.
The structural elements similar between kinesin and myosin are not contiguous, but are interrupted by large amino acid insertions. Myosin has two major insertions that correspond to much smaller loops in kinesin: a ~180 aa domain between residues 270 and 450 (insertion 1) which corresponds to a smaller insertion in kinesin (aa 138-173), and a 142 aa domain between residues 506 and 649 (insertion 2) which corresponds to loop L12 in kinesin (aa 272-280). Myosin also has longer N- and C-terminal extensions. The structural similarity between the kinesin and myosin cores suggests that these motors diverged from a common ancestor and may use a similar strategy for converting chemical energy into mechanical work.
Comparison of Kinesins to G proteins
The architecture and residues in the nucleotide pocket of kinesin and Ncd show similarity to G proteins. Like motor proteins, G proteins undergo a nucleotide hydrolysis cycle which is coupled to conformational changes that alter binding to other macromolecules. Therefore, G proteins could potentially provide insight into force transduction in motor proteins.
Kinesin motors and G proteins do not have extended structural or sequence similarity. However, four conserved sequence motifs in the kinesin, myosin and G protein families are located in equivalent positions. In G proteins, these regions interact with nucleotide and are termed G-1 (P-loop), G-2 (D-(x)n-T), G-3 (DxxG) and G-4 (N/TKxD). The G-2 region is in an equivalent position to the N-2 region of motor proteins (NxxSSR). The G-2 threonine is located in a position similar to a N-2 serine. The G-3 region corresponds to N-3 in motors (DxxGxE). Notably, the side chains of the Asp and the C-alpha of the glycine residues are in very similar locations in p21ras and Ncd. The G-4 region is also located in a position similar to N-4 in kinesin proteins (RxRP) and NP for myosins. The lysine of G-4 and the second arginine of N-4 superimpose and both form contacts with the purine ring of nucleotide. The structural comparisons of Ncd, kinesin, myosin and G proteins suggest that these NTPases may utilize a similar strategy of changing conformation between NTP and NDP states.
Contributed by Ron Vale
References
Kull, F.J., Sablin, E.P., Lau, R., Fletterick, R.J. and Vale, R.D. 1996. Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature 380:550-555.Sablin, E.P., Kull, F.J., Cooke, R., Vale, R.D. and Fletterick, R.J. 1996. Crystal structure of the motor domain of the kinesin-related motor ncd. Nature 380:555-559.Vale, R.D. 1996. Switches, latches, and levers: common themes of G proteins and molecular motors. J. Cell Biol 135(2):291-302
View conserved motifs positioned on the kinesin motor domain.
View link to Simulated Annealing of Kinesin.
View conditions used to crystallize kinesin proteins.View links to recent articles on kinesins and crystal structure from the MEDLINE database.Go to the Domain Organization & Conformational Changes page.
Go to the Kinesin Interactions with Microtubules page.
Return to the Kinesin Home Page.If you use material from the reviews at the Kinesin site, we ask that you cite either the home page and author, or the appropriate source publication in your work.Copyright 1996, 1997, 1998 All rights reservedCreated 19 August 1996 20:00 GMT
Modified 1 July 1999 19:01 GMT
Kinesin Structures
Tubulin/MT StructuresCoordinates |