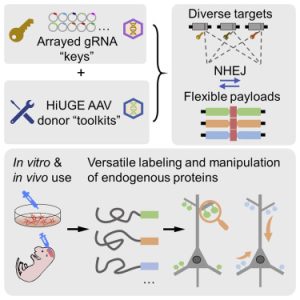
One of the focuses of the Soderling Lab is the mechanisms underlying disrupted synaptic connections, abnormalities that play a role in the onset of neurodegenerative and psychiatric disorders. To accomplish this, the lab has developed a combination of proteomic and CRISPR approaches, such as the CRISPR-Cas9-based Homology-independent Universal Genome Engineering (HiUGE) method. HiUGE allows efficient and convenient alteration of endogenous proteins through adeno-associated virus (AAV) vectors of insertional DNA sequences that can merge into the part of the genome specified by guide-RNA vectors (Gao et al., 2019). As demonstrated in the following Graphical Abstract from Gao et al., HiUGE provides a simplified way to modify proteins in vivo and in vitro for the study of gene and proteins functions, thus making it easier to investigate the proteins and synaptic disruptions involved in diseases such as Parkinson’s (2019).
Parkinson’s disease (PD) is a progressive, age-associated neurodegenerative movement disorder often characterized by a resting tremor, slow movement, and difficulty maintaining posture due to the loss of dopamine-producing neurons in the midbrain. The etiology of the disease continues to be studied, with advances in the identification of gene mutations specific to familial PD encouraging further inquiry into the mechanisms by which these mutations induce it. One such PD-linked gene is the vacuolar protein sorting 35 ortholog (VPS35), with the VPS35 D620N mutation having been identified as pathogenic (Williams et al., 2017).
Together, HiUGE and PD are the foundation for my research project this summer. I will be investigating the factors that may improve the efficacy of the former while simultaneously employing HiUGE to observe the latter in vitro (and possibly in vivo). Currently, I have three research questions that include the following:
- Are dual-oriented HiUGE-donors more efficient than single-oriented ones for gene expression?
- What is the effect of knock in WT/DG20N VPS35 – a dual-oriented HiUGE-donor – in vitro (and potentially in vivo)?
- Does UltraID produce biotinylation of alpha synuclein comparable to TurboID?
The first question relates to whether a DNA vector or construct created through the HiUGE method might produce double the expression of a specific gene if its epitope is dual-oriented versus single-oriented. The second question involves a dual-oriented HiUGE donor with healthy and PD-linked DNA fragments being introduced in vitro to a cell culture and potentially in vivo with mouse models. This second question also involves partially testing the efficacy of the dual-oriented HiUGE donor. Lastly, the third question has ramifications as to whether labeling proteins such as alpha synuclein and others potentially linked to PD in the synapses can be equally if not more effective through the use of the enzyme UltraID, a smaller molecule as opposed to the currently used TurboID.
I look forward to continuing to learn as I seek the answers to these and other questions in the Soderling Lab this summer.


 Graphical Abstract from
Graphical Abstract from