Research
Research Highlights
Reviews and Foundational Concepts
Note: The image of each paper's title contains a hyperlink to the publication.
Metabolomics - A Biochemical Approach for Understanding Drug Response
Metabolomics is the study of metabolism at the global level. This rapidly developing new discipline has important potential implications for pharmacologic science. The concept that metabolic state is representative of the overall physiologic status of the organism lies at the heart of metabolomics. Metabolomic studies capture global biochemical events by assaying thousands of small molecules in cells, tissues, organs, or biological fluids—followed by the application of informatic techniques to define metabolomic signatures. Metabolomic studies can lead to enhanced understanding of disease mechanisms and to new diagnostic markers as well as enhanced understanding of mechanisms for drug or xenobiotic effect and increased ability to predict individual variation in drug response phenotypes (pharmacometabolomics). This review outlines the conceptual basis for metabolomics as well as analytical and informatic techniques used to study the metabolome and to define metabolomic signatures. It also highlights potential metabolomic applications to pharmacology and clinical pharmacology.
Metabolomics for the study of CNS diseases
Metabolomics, the omics science of biochemistry, is a global approach to understanding regulation of metabolic pathways and metabolic networks of a biological system. Metabolomics complements data derived from genomics, transcriptomics, and proteomics to assist in providing a systems approach to the study of human health and disease. In this review we focus on applications of metabolomics for the study of diseases of the nervous system. We share concepts in metabolomics, tools used in metabolic profiling and early findings from the study of neuropsychiatric diseases, and drugs used to treat these diseases. Metabolomics emerges as another powerful tool in central nervous system research.
Pharmacometabolomics Enables Precision Medicine
The scaling up of data in clinical pharmacology and the merger of systems biology and pharmacology has led to the emergence of a new discipline of Quantitative and Systems Pharmacology (QSP). This new research direction might significantly advance the discovery, development, and clinical use of therapeutic drugs. Research communities from computational biology, systems biology, and biological engineering--working collaboratively with pharmacologists, geneticists, biochemists, and analytical chemists--are creating and modeling large data on drug effects that is transforming our understanding of how these drugs work at a network level. In this review, we highlight developments in a new and rapidly growing field--pharmacometabolomics--in which large biochemical data-capturing effects of genome, gut microbiome, and environment exposures is revealing information about metabotypes and treatment outcomes, and creating metabolic signatures as new potential biomarkers. Pharmacometabolomics informs and complements pharmacogenomics and together they provide building blocks for QSP.
Key Research Papers
Pharmacometablomics-informs pharmacogenomics

Manhattan plot for baseline plasma KYN concentrations. SNPs across the DEFB1 and AHR genes have been highlighted, with rs5743467 as the “top” DEFB1 SNP (P-value = 8.18E−07) and, rs17137566 as the “top” AHR SNP (P-value = 6.22E−06).
Major depressive disorder (MDD) is a heterogeneous disease. Efforts to identify biomarkers for sub-classifying MDD and antidepressant therapy by genome-wide association studies (GWAS) alone have generally yielded disappointing results. We applied a metabolomics-informed genomic research strategy to study the contribution of genetic variation to MDD pathophysiology by assaying 31 metabolites, including compounds from the tryptophan, tyrosine, and purine pathways, in plasma samples from 290 MDD patients. Associations of metabolite concentrations with depressive symptoms were determined, followed by GWAS for selected metabolites and functional validation studies of the genes identified. Kynurenine (KYN), the baseline plasma metabolite that was most highly associated with depressive symptoms, was negatively correlated with severity of those symptoms. GWAS for baseline plasma KYN concentrations identified SNPs across the beta-defensin 1 (DEFB1) and aryl hydrocarbon receptor (AHR) genes that were cis-expression quantitative trait loci (eQTLs) for DEFB1 and AHR mRNA expression, respectively. Furthermore, the DEFB1 locus was associated with severity of MDD symptoms in a larger cohort of 803 MDD patients. Functional studies demonstrated that DEFB1 could neutralize lipopolysaccharide-stimulated expression of KYN-biosynthesizing enzymes in monocytic cells, resulting in altered KYN concentrations in the culture media. In addition, we demonstrated that AHR was involved in regulating the expression of enzymes in the KYN pathway and altered KYN biosynthesis in cell lines of hepatocyte and astrocyte origin. In conclusion, these studies identified SNPs that were cis-eQTLs for DEFB1 and AHR and, which were associated with variation in plasma KYN concentrations that were related to severity of MDD symptoms.
Response to sertraline
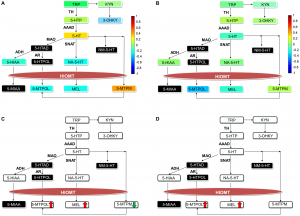
Panels A and B show correlations of metabolites at baseline with the four week treatment response in the sertraline and placebo groups, respectively. The correlations are color coded as indicated by the color bar. Panels C and D show four week metabolic changes unique to responders in the sertraline and placebo groups respectively. Abbreviations: HIOMT = hydroxyindole-O-methyltransferase; 5-HIAD = 5-Hydroxyindole acetyldehyde; NM-5-HT = N-methyl serotonin; 5-HTPOL = 5-Hydroxytryptophol; 5-MIAA = 5-Methoxyindole acetic acid; 5-MTPM = 5-methoxytryptamine.
Therapeutic response to selective serotonin (5-HT) reuptake inhibitors in Major Depressive Disorder (MDD) varies considerably among patients, and the onset of antidepressant therapeutic action is delayed until after 2 to 4 weeks of treatment. The objective of this study was to analyze changes within methoxyindole and kynurenine (KYN) branches of tryptophan pathway to determine whether differential regulation within these branches may contribute to mechanism of variation in response to treatment. Metabolomics approach was used to characterize early biochemical changes in tryptophan pathway and correlated biochemical changes with treatment outcome. Outpatients with MDD were randomly assigned to sertraline (n = 35) or placebo (n = 40) in a double-blind 4-week trial; response to treatment was measured using the 17-item Hamilton Rating Scale for Depression (HAMD17). Targeted electrochemistry based metabolomic platform (LCECA) was used to profile serum samples from MDD patients. The response rate was slightly higher for sertraline than for placebo (21/35 [60%] vs. 20/40 [50%], respectively, χ2(1) = 0.75, p = 0.39). Patients showing a good response to sertraline had higher pretreatment levels of 5-methoxytryptamine (5-MTPM), greater reduction in 5-MTPM levels after treatment, an increase in 5-Methoxytryptophol (5-MTPOL) and Melatonin (MEL) levels, and decreases in the (KYN)/MEL and 3-Hydroxykynurenine (3-OHKY)/MEL ratios post-treatment compared to pretreatment. These changes were not seen in the patients showing poor response to sertraline. In the placebo group, more favorable treatment outcome was associated with increases in 5-MTPOL and MEL levels and significant decreases in the KYN/MEL and 3-OHKY/MEL; changes in 5-MTPM levels were not associated with the 4-week response. These results suggest that recovery from a depressed state due to treatment with drug or with placebo could be associated with preferential utilization of serotonin for production of melatonin and 5-MTPOL.
Potential predictors of response to treatment

Partial least square-discriminant analysis (PLS-DA) scores plot of responders and non-responders on sertraline
The purpose of this study was to determine whether the baseline metabolic profile (that is, metabotype) of a patient with major depressive disorder (MDD) would define how an individual will respond to treatment. Outpatients with MDD were randomly assigned to sertraline (up to 150 mg per day) (N=43) or placebo (N=46) in a double-blind 4-week trial. Baseline serum samples were profiled using the liquid chromatography electrochemical array; the output was digitized to create a ‘digital map’ of the entire measurable response for a particular sample. Response was defined as ⩾50% reduction baseline to week 4 in the 17-item Hamilton Rating Scale for Depression total score. Models were built using the one-out method for cross-validation. Multivariate analyses showed that metabolic profiles partially separated responders and non-responders to sertraline or to placebo. For the sertraline models, the overall correct classification rate was 81% whereas it was 72% for the placebo models. Several pathways were implicated in separation of responders and non-responders on sertraline and on placebo including phenylalanine, tryptophan, purine and tocopherol. Dihydroxyphenylacetic acid, tocopherols and serotonin were common metabolites in separating responders and non-responders to both drug and placebo. Pretreatment metabotypes may predict which depressed patients will respond to acute treatment (4 weeks) with sertraline or placebo. Some pathways were informative for both treatments whereas other pathways were unique in predicting response to either sertraline or placebo. Metabolomics may inform the biochemical basis for the early efficacy of sertraline.
Recent Representative Papers
Translational Psychiatry, 04 July 2019

Metabolite changes associated with HRSD17 scores.
Metabolomics provides valuable tools for the study of drug effects, unraveling the mechanism of action and variation in response due to treatment. In this study we used electrochemistry-based targeted metabolomics to gain insights into the mechanisms of action of escitalopram/citalopram focusing on a set of 31 metabolites from neurotransmitter-related pathways. Overall, 290 unipolar patients with major depressive disorder were profiled at baseline, after 4 and 8 weeks of drug treatment. The 17-item Hamilton Depression Rating Scale (HRSD17) scores gauged depressive symptom severity. More significant metabolic changes were found after 8 weeks than 4 weeks post baseline. Within the tryptophan pathway, we noted significant reductions in serotonin (5HT) and increases in indoles that are known to be influenced by human gut microbial cometabolism. 5HT, 5-hydroxyindoleacetate (5HIAA), and the ratio of 5HIAA/5HT showed significant correlations to temporal changes in HRSD17 scores. In the tyrosine pathway, changes were observed in the end products of the catecholamines, 3-methoxy-4-hydroxyphenylethyleneglycol and vinylmandelic acid. Furthermore, two phenolic acids, 4-hydroxyphenylacetic acid and 4-hydroxybenzoic acid, produced through noncanconical pathways, were increased with drug exposure. In the purine pathway, significant reductions in hypoxanthine and xanthine levels were observed. Examination of metabolite interactions through differential partial correlation networks revealed changes in guanosine–homogentisic acid and methionine–tyrosine interactions associated with HRSD17. Genetic association studies using the ratios of these interacting pairs of metabolites highlighted two genetic loci harboring genes previously linked to depression, neurotransmission, or neurodegeneration. Overall, exposure to escitalopram/citalopram results in shifts in metabolism through noncanonical pathways, which suggest possible roles for the gut microbiome, oxidative stress, and inflammation-related mechanisms.
Frontiers Neuroscience, 12 Sept. 2019

The pairwise correlations between the changes in the metabolite members to each other and also to HAM-D17 changes are shown in a composite plot.
Major depressive disorder (MDD) is a common and disabling syndrome with multiple etiologies that is defined by clinically elicited signs and symptoms. In hopes of developing a list of candidate biological measures that reflect and relate closely to the severity of depressive symptoms, so-called “state-dependent” biomarkers of depression, this pilot study explored the biochemical underpinnings of treatment response to cognitive behavior therapy (CBT) in medication-free MDD outpatients. Plasma samples were collected at baseline and week 12 from a subset of MDD patients (N = 26) who completed a course of CBT treatment as part of the Predictors of Remission in Depression to Individual and Combined Treatments (PReDICT) study. Targeted metabolomic profiling using the AbsoluteIDQ® p180 Kit and LC-MS identified eight “co-expressed” metabolomic modules. Of these eight, three were significantly associated with change in depressive symptoms over the course of the 12-weeks. Metabolites found to be most strongly correlated with change in depressive symptoms were branched chain amino acids, acylcarnitines, methionine sulfoxide, and α-aminoadipic acid (negative correlations with symptom change) as well as several lipids, particularly the phosphatidlylcholines (positive correlation). These results implicate disturbed bioenergetics as an important state marker in the pathobiology of MDD. Exploratory analyses contrasting remitters to CBT versus those who failed the treatment further suggest these metabolites may serve as mediators of recovery during CBT treatment. Larger studies examining metabolomic change patterns in patients treated with pharmacotherapy or psychotherapy will be necessary to elucidate the biological underpinnings of MDD and the -specific biologies of treatment response.
Journal Affective Disorders, 1 Oct. 2018

The heatmap depicts the log2 fold change of metabolite levels from baseline to week eight of SSRI treatment. CD+: Core Depression, ANX+: Anxiety, NVSM+: Neurovegetative Symptom of Melancholia. P-values were obtained using linear mixed effect models controlling for age, sex and baseline 17-item Hamilton Rating Scale for Depression scores. Red indicates an increase and blue indicates a decrease in metabolite levels over eight weeks of treatment; *:p-value<0.05, **:p-value<0.01 and ***:p-value<0.001.
Metabolic Signature of Eight Weeks of Exposure to SSRI.
- This study was to assess whether three symptomatically defined phenotypes of MDD, (core depression, neurovegetative of melancholia and anxiety), could be differentiated based on acylcarnitine profiles at baseline, after eight weeks of citalopram/escitalopram treatment.
- The current data demonstrated that these phenotypes have distinct patterns of acylcarnitine levels at baseline and after eight weeks of antidepressant treatment.
- These findings may help to develop a metabolomic profile of MDD patients with the aim of improving subtype classification of the MDD syndrome.









