The exon junction complex: canonical and non-canonical control of brain development

The RNA binding exon junction complex (EJC), composed of Magoh, Rbm8a and Eif4a3, is indispensible for mRNA fate and function throughout eukaryotes. Mutations and copy number variations in EJC components are associated with human microcephaly, autism and intellectual disability. See this review on the EJC and this review on RNA binding proteins to learn more. Over the years we have discovered that haploinsufficiency for EJC components, causes microcephaly, due to defects in neural progenitor proliferation and neuronal apoptosis (Nature Neuroscience and The Journal of Neuroscience). Genetics, transcriptomics and proteomics have demonstrated which components control cortical development (RNA), and revealed common alterations downstream of EJC dysfunction (PLoS Genetics)! We have collaborated with the Passo-Bueno lab to model how EIF4A3 mutations disrupt neural crest to cause Richieri-Costa-Peirera Syndrome (RCPS) (Hum Mol Genetics). Our work reveals disease relevant roles for EJC components in brain development.
Studies of the EJC along with pharmacology and live imaging have led us to discover new links between mitosis length of stem cells and cell fate. We discovered that Magoh haploinsufficient neural progenitors exhibit mitotic delay, and these progenitors directly produce more neurons instead of new progenitors. This fascinating phenotype is recapitulated using pharmacology-in vitro, ex vivo and in vivo, revealing that prolonged progenitor mitosis is sufficient to alter neural cell fates and identifying one explanation for microcephaly (Neuron and Developmental Neuroscience). We have extended these findings to interneuron development and human neural progenitors. We discovered that Magoh is essential for interneuron generation and survival linked to prolonged mitosis of interneuron progenitors (Development). Our study in Development used fixed and live imaging of mouse and human neural progenitors to demonstrate critical roles for Eif4a3 in mitosis duration of both species.
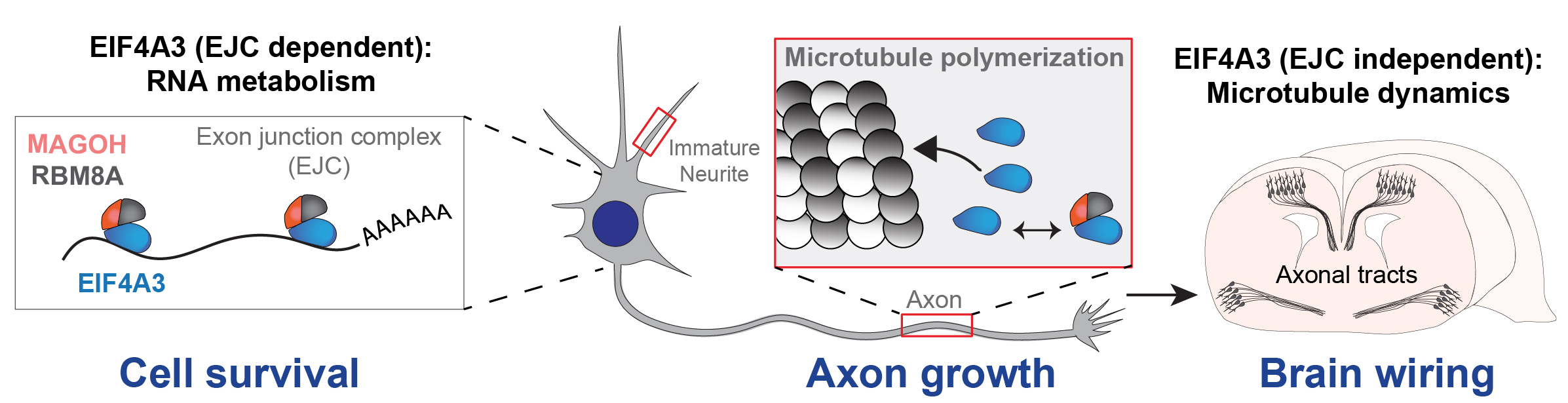 Does the EJC only control RNA? Our recent study in Cell Reports defines a new non-canonical role for EIF4A3 in axon formation in vivo, by directly binding to microtubules and controlling their dynamics. This study uses mouse genetics, patient derived cortical organoids, live imaging, and protein biochemistry to define these new functions. Our study reveals a fundamental mechanism by which neurons re-utilize core gene expression machinery to directly control the cytoskeleton. This challenges the notion that RNA-binding proteins exclusively control RNA metabolism in the nervous system.
Does the EJC only control RNA? Our recent study in Cell Reports defines a new non-canonical role for EIF4A3 in axon formation in vivo, by directly binding to microtubules and controlling their dynamics. This study uses mouse genetics, patient derived cortical organoids, live imaging, and protein biochemistry to define these new functions. Our study reveals a fundamental mechanism by which neurons re-utilize core gene expression machinery to directly control the cytoskeleton. This challenges the notion that RNA-binding proteins exclusively control RNA metabolism in the nervous system.
We continue to use mouse and human models to understand how the EJC controls brain development and disease and how it controls microtubules.
DDX3X and DDX3X syndrome: translational control of neurogenesis
 We investigate the RNA binding protein DDX3X to better understand how the spectrum of mutations in this essential gene cause disease. De novo mutations in DDX3X underlie 1-3% of female intellectual disabilities and cause both DDX3X Syndrome and Autism. We aim to understand how the diverse landscape of DDX3X missense and nonsense mutations affect brain development. In previous collaborations with Dr. Elliott Sherr and Stephen Floor we discovered that depletion of DDX3X impairs neuron number by disrupting progenitors. Further we showed that missense mutations in DDX3X cause abnormal RNA-protein granule formation in progenitors. This work was published in 2020 in Neuron. Our eLife study phenotypes a loss of function Ddx3x mouse model, informing new requirements for DDX3X in progenitor cell cycle, as well as using ribosome profiling to discover DDX3X translational targets in the brain. Our recent study in PLOS Genetics uses live imaging, transcriptomics, and proteomics to discover how clinically diverse missense mutations in DDX3X influence neural cell fate. Our findings reveal new mechanisms by which clinically distinct DDX3X missense mutations differentially impair neurodevelopment.
We investigate the RNA binding protein DDX3X to better understand how the spectrum of mutations in this essential gene cause disease. De novo mutations in DDX3X underlie 1-3% of female intellectual disabilities and cause both DDX3X Syndrome and Autism. We aim to understand how the diverse landscape of DDX3X missense and nonsense mutations affect brain development. In previous collaborations with Dr. Elliott Sherr and Stephen Floor we discovered that depletion of DDX3X impairs neuron number by disrupting progenitors. Further we showed that missense mutations in DDX3X cause abnormal RNA-protein granule formation in progenitors. This work was published in 2020 in Neuron. Our eLife study phenotypes a loss of function Ddx3x mouse model, informing new requirements for DDX3X in progenitor cell cycle, as well as using ribosome profiling to discover DDX3X translational targets in the brain. Our recent study in PLOS Genetics uses live imaging, transcriptomics, and proteomics to discover how clinically diverse missense mutations in DDX3X influence neural cell fate. Our findings reveal new mechanisms by which clinically distinct DDX3X missense mutations differentially impair neurodevelopment.
We continue to use mouse models, cell based models, genomics and imaging to understand how DDX3X mutations impair brain development.
Control of RNA stability in cortical development
RNA abundance is controlled by rates of synthesis and degradation. Although mis-regulation of RNA turnover is linked to neurodevelopmental disorders, how it contributes to cortical development is largely unknown. Our recent work has used SLAM-seq to discover the landscape of RNA stability regulation in the cerebral cortex. We discovered new features controlling RNA stability and identify a link between RNA half life and developmental expression. Further, using compound mouse genetics, we discover CNOT3, a core component of the CCR4-NOT deadenylase complex linked to neurodevelopmental disease, is essential for cortical development. Collectively, our findings demonstrate that fine-tuned control of RNA turnover is crucial for brain development. This work was published in PLOS Biology.
We continue to use omics approaches to gain an integrated understanding of how the transcriptome is shaped post-transcriptionally to control cortical development.