Microbial regulation of host nutrient metabolism and immunity
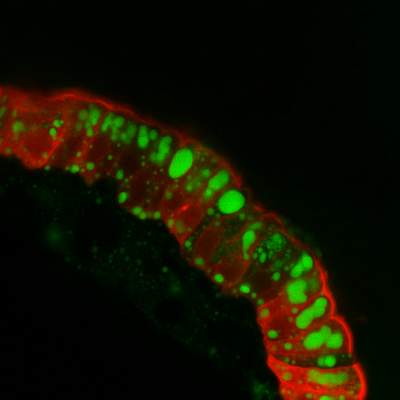 Digestion and absorption of dietary nutrients are the primary physiologic functions of the intestinal tract. Impairments and augmentations in dietary nutrient harvest contribute respectively to malnutrition and cardiometabolic disease. Despite the central importance of intestinal nutrient assimilation in metabolic health, our understanding of underlying cellular and molecular mechanisms is far from complete. The ability of the microbiome to modify dietary nutrient metabolism has emerged as a key feature of host-microbe relationships in the gut. This capacity has been most extensively described in the mammalian colon where microbial fermentation of otherwise indigestible complex carbohydrates in the diet produces monosaccharides and short-chain fatty acids that can then be absorbed by the host. However, the vast majority of dietary nutrient digestion and absorption occurs in the small intestine where the contributions of the microbiome remained relatively unexplored.
Digestion and absorption of dietary nutrients are the primary physiologic functions of the intestinal tract. Impairments and augmentations in dietary nutrient harvest contribute respectively to malnutrition and cardiometabolic disease. Despite the central importance of intestinal nutrient assimilation in metabolic health, our understanding of underlying cellular and molecular mechanisms is far from complete. The ability of the microbiome to modify dietary nutrient metabolism has emerged as a key feature of host-microbe relationships in the gut. This capacity has been most extensively described in the mammalian colon where microbial fermentation of otherwise indigestible complex carbohydrates in the diet produces monosaccharides and short-chain fatty acids that can then be absorbed by the host. However, the vast majority of dietary nutrient digestion and absorption occurs in the small intestine where the contributions of the microbiome remained relatively unexplored.
Our lab has made pioneering contributions to our understanding of how the microbiome regulates dietary nutrient metabolism in the small intestine and responds to nutrient availability. We first made the landmark discovery that the microbiome stimulates IEC absorption of dietary fats and their subsequent metabolism in the liver. We also recently established the zebrafish as a model for studying physiology and environmental regulation of enteroendocrine cells (EECs). EECs are specialized sensory epithelial cells that detect luminal nutrients and microbial products, and communicate that information to the rest of the body via release of hormones and direct synaptic connections with sensory neurons. Using a combination of in vivo imaging, genetic and gnotobiotic manipulations in zebrafish, we are exploring how EECs respond and adapt to nutritional and microbial stimuli, and how these EEC responses impact upon systemic metabolism and gut-brain communication. We are currently seeking to define the specific molecular mechanisms by which bacteria regulate these aspects of host metabolic physiology. To this end, we are establishing genetic methods in bacterial strains known to regulate zebrafish metabolism such as Exiguobacterium acetylicum. We are also working with collaborators to develop organoid platforms for analysis of host-microbe interactions and nutrient metabolism in the mammalian intestine.
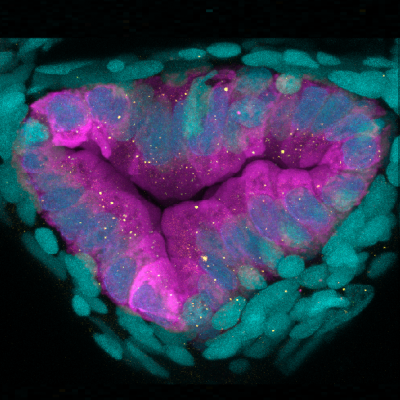 The intestine faces the daunting challenge of maintaining homeostasis in the context of dynamic microbial and chemical challenges. Impairment in these physiologic functions can contribute to the etiology of sepsis, IBD, and other inflammatory diseases. The mechanisms by which the host maintains intestinal barrier function and homeostasis, and modulates the immune system in response to these challenges, remain unresolved. Chemical models of intestinal injury in mice (e.g., DSS, TNBS, Oxazolone, NSAIDs) have proven to be useful tools for identifying mechanisms underlying epithelial barrier function, inflammation, and restitution. Our work has established the NSAID Glafenine as a useful chemical model of intestinal injury in the zebrafish, and used it to uncover protective roles for multi-drug resistance efflux pumps and IEC shedding in the maintenance of intestinal homeostasis. Our work has also advanced understanding of the host effector proteins that communicate microbiome colonization status to the systemic innate immune system. We found that microbiome colonization affects diverse aspects of intestinal and systemic neutrophil function. We also recently discovered that a host effector protein produced by the intestine in response to microbiome called Serum amyloid A (Saa) serves as a systemic signal to neutrophils to restrict aberrant activation, decreasing inflammatory tone and bacterial killing potential while simultaneously enhancing their ability to migrate to wounds. This work provides a critical frame of reference for our current efforts to define novel immunoregulatory factors produced by the intestine and other tissues in response to members of the microbiome.
The intestine faces the daunting challenge of maintaining homeostasis in the context of dynamic microbial and chemical challenges. Impairment in these physiologic functions can contribute to the etiology of sepsis, IBD, and other inflammatory diseases. The mechanisms by which the host maintains intestinal barrier function and homeostasis, and modulates the immune system in response to these challenges, remain unresolved. Chemical models of intestinal injury in mice (e.g., DSS, TNBS, Oxazolone, NSAIDs) have proven to be useful tools for identifying mechanisms underlying epithelial barrier function, inflammation, and restitution. Our work has established the NSAID Glafenine as a useful chemical model of intestinal injury in the zebrafish, and used it to uncover protective roles for multi-drug resistance efflux pumps and IEC shedding in the maintenance of intestinal homeostasis. Our work has also advanced understanding of the host effector proteins that communicate microbiome colonization status to the systemic innate immune system. We found that microbiome colonization affects diverse aspects of intestinal and systemic neutrophil function. We also recently discovered that a host effector protein produced by the intestine in response to microbiome called Serum amyloid A (Saa) serves as a systemic signal to neutrophils to restrict aberrant activation, decreasing inflammatory tone and bacterial killing potential while simultaneously enhancing their ability to migrate to wounds. This work provides a critical frame of reference for our current efforts to define novel immunoregulatory factors produced by the intestine and other tissues in response to members of the microbiome.
Relevant publications:
Kanther, M., Sun. S., Mühlbauer, M., Mackey, L.C., Flynn, E.J., Bagnat, M., Jobin, C., and Rawls, J.F. (2011) Microbial colonization induces dynamic temporal and spatial NF-κB responses in the zebrafish digestive tract. Gastroenterology 141(1): 197-207. [PMCID: PMC3164861]
Semova, I., Carten, J.D., Stombaugh, J., Mackey, L.C., Knight, R., Farber, S.A., and Rawls, J.F. (2012) Microbiota and diet regulate fatty acid absorption and metabolism in the zebrafish intestine. Cell Host & Microbe 12: 277–288. [PMCID: PMC3517662]
Kanther, M., Tomkovich, S.1, Sun, X., Grosser, M.R., Koo, J., Flynn, E.J., Jobin, C., and Rawls, J.F. (2014) Commensal microbiota stimulate systemic neutrophil migration through induction of Serum amyloid A. Cell. Microbiol. 16(7): 1053-67.
[PMCID: PMC4364439]
Goldsmith, J.R., Cocchiaro, J.L., Rawls, J.F., and Jobin, C. (2013) Glafenine-intestinal injury in zebrafish is ameliorated by mu-opioid signaling via enhancement of Atf6-dependent cellular stress responses. Dis. Models & Mech. 6(1):146-159.
[PMCID: PMC3529347]
Wong, S., Burns, A.R., Stephens, W.Z., Stagaman, K., David, L.A., Guillemin, K., and Bohannan, B.J.M., and Rawls, J.F. (2015) Ontogenetic differences in dietary fat influence microbiota assembly in the zebrafish gut. MBio 6(5): e00687-15.
[PMCID: PMC4611033]
Bae, S., Mueller, O., Wong, S., Rawls, J.F., and Valdivia, R.H. (2016) Genomic sequencing-based mutational enrichment analysis identifies motility genes in a genetically intractable gut microbe. Proc Natl Acad Sci USA 113(49): 14127-14132. [PMCID: PMC5150387]
Williamson, I.A., Arnold, J.W., Samsa, L.A., Gaynor, L. DeSalvo, M., Cocchiaro, J.L., Carroll, I., Azcarate-Peril, A., Rawls, J.F., Allbritton, N.L., and Magness, S.T. (2018) A high-throughput organoid microinjection platform to study gastrointestinal luminal physiology and the microbiome. Cellular and Molecular Gastroenterology and Hepatology 6(3):301-319. [PMCID: PMC6092482]
Murdoch, C.C., Espenschied, S.T., Matty, M.A., Mueller, O., Tobin, D.M. and Rawls, J.F. (2019) Microbiota-induced Serum Amyloid A in the intestine directs systemic neutrophil function. PLoS Pathogens (in press)
Espenschied, S.T., Wen, J., Cronan, M.R., Matty, M.A., Mueller, O., Redinbo, M.R., Tobin, D.M., and Rawls, J.F. Epithelial delamination is protective during pharmaceutical-induced enteropathy. (submitted)