The Poss lab focuses on the concepts and mechanisms of regeneration in zebrafish. Zebrafish are a known animal model system for studying regeneration because of their remarkable ability to regenerate lost tissue after injury. Adult zebrafish are capable of regenerating tissue in the heart, as well as fin, spinal cord, and retina.
Poss, Kenneth D. “Getting to the Heart of Regeneration in Zebrafish.” Seminars in Cell & Developmental Biology 18.1 (2007): 36-45.
In fact, it has been shown that zebrafish are able to regenerate their hearts, not due to stem cells, but the proliferation of spared cardiomyocytes (Poss, Wilson, & Keating 2002).
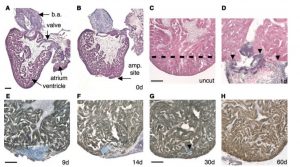
Poss, Kenneth D., Lindsay G. Wilson, and Mark T. Keating. “Heart Regeneration in Zebrafish.” Science 298.5601 (2002): 2188-190. 1 July 2016.
This picture shows the regeneration of ventricular myocardium in the resected zebrafish heart. After 20% of the ventricle was amputated off, by day 60, no sign of injury remained and the heart is fully repaired.
In contrast, mammals do not have this capacity to regenerate and repair its tissue efficiently.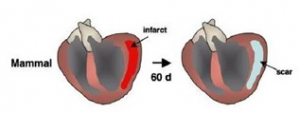
Poss, Kenneth D. “Getting to the Heart of Regeneration in Zebrafish.” Seminars in Cell & Developmental Biology 18.1 (2007): 36-45.
Thus, injury in the heart often leads to scarring and fibrosis. This makes diseases such as heart disease a significant threat to human health. Studying the mechanisms of regeneration in zebrafish can illuminate factors that may be applicable to the stimulation of human heart regeneration.
My research project focuses on heart development. Specifically, I am looking at 4 different genes and the roles they play in cardiac development and regeneration. The zebrafish heart is divided into two main parts – the atrium and the ventricle. The overall concept of this project is to examine what factors in heart development are responsible in pushing some cells into differentiating into the atrium while pushing other cells into becoming the ventricle. In order to examine these genes’ influence in heart development, I look at whether these 4 genes are atrial specific, meaning they are uniquely expressed in the atrium, or whether there are ventricle specific, meaning they are only present in the ventricle. Two of the genes I am examining were hypothesized to be atrial specific while the other two genes were hypothesized to be ventricle specific.
In order to examine where in the heart these genes are being expressed, I am performing in situ hybridization with adult zebrafish hearts. In situ hybridization is a method that uses a labeled complementary piece of DNA or RNA to locate a segment of DNA or RNA of the gene of interest in tissue (Wikipedia). I will first make a RNA probe that is a segment of RNA complementary to one of the genes of interest. This RNA probe will contain digoxygenin tags that will then be used to immunohistochemically label cells expressing the complementary mRNA. The areas of the heart where the gene is being actively expressed will turn blue. With this method, I will be able to see exactly where in the heart the genes of interest are being expressed. I will also perform in situ hybridization on 6 week old zebrafish to see where the genes are active during the earlier stages of zebrafish life. Furthermore, I will perform in situ hybridization on injured adult zebrafish hearts in order to see where in the heart these genes are being expressed during regeneration and if there are any changes in levels of expression compared to uninjured adult zebrafish hearts. The next step in this project would be to identify possible atrial or ventricle specific enhancers of these genes, clone them in front of a GFP gene, inject into zebrafish embryos, and observe where in the heart these enhancers are active (turns green). Then, CRISPR/Cas9 gene editing would be used to delete the enhancers of these genes and to examine the effects of the deletion on the cardiac development of zebrafish. Overall, with this project I am hoping to find out what roles these four specific genes play in heart development and regeneration.
Unlike the majority of the Poss Lab that focuses on regeneration, my project looks more at heart development. However, development and regeneration are closely linked, so I hope I will be able to contribute information on the roles of these genes in the heart in terms of both cardiac development and regeneration.




