Our lab aims to gain a mechanistic understanding of biology at the molecular level. We integrate biochemical and biophysical (NMR, crystallography, and cryo-EM) tools to investigate enzymes and molecular machineries as well as their regulation and inhibition by small molecules. Insights gained from our studies have enabled the discovery of new enzymatic functions and signaling pathways, development of novel antibiotics and cancer therapeutics, and engineering of disease-resistant crops and crop-protection chemicals.
1. Gram-negative bacterial outer membrane biogenesis
Our lab has systematically characterized bacterial enzymes involved in the biosynthesis and modification of lipid A, the membrane anchor of lipopolysaccharide (LPS) / lipooligosaccharide (LOS) and the major lipid component of the outer leaflet of the Gram-negative bacterial outer membrane. These studies have contributed to a molecular understanding of the structure and mechanism of lipid A enzymes, and have led to the development of novel antibiotics targeting LpxC and LpxH. Our lead LpxC inhibitor, LPC-233, has demonstrated extraordinary in vitro and in vivo antibiotic activities and safety and has been licensed by Valanbio Therapeutics for clinical development of novel antibiotics.

• Design and Evaluation of Pyridinyl Sulfonyl Piperazine LpxH Inhibitors with Potent Antibiotic Activity Against Enterobacterales. Ennis AF†, Cochrane CS†, Dome PA†, Jeong P, Yu J, Lee H, Williams CS, Ha Y, Yang W, Zhou P* and Hong J*. JACS Au 2024; 4, 4383-4393.
• Preclinical safety and efficacy characterization of an LpxC inhibitor against Gram-negative pathogens. Zhao J, Cochrane CS, Najeeb J, Gooden D, Sciandra C, Fan P, Lemaitre N, Newns K, Nicholas RA, Guan Z, Thaden JT, Fowler VG Jr, Spasojevic I, Sebbane F, Toone EJ, Duncan C, Gammans R, Zhou P*. Sci Transl Med. 2023; 15: eadf5668.

2. Host-microbe interactions
Leveraging structural biology and molecular modeling tools, our lab has elucidated how bacterial pathogens manipulate host/plant cells through their injected effector proteins, such as AvrE/DspE (Nature 2023) and how plant cells counter pathogenic infections through systemic acquired resistance (SAR) mediated by NPR1 (Nature 2022). Our work, carried out in collaboration with Drs. Sheng-Yang He and Ke Dong (Duke Biology), has demonstrated that AvrE/DspE proteins form bacterial porin-like channels and directly transport a variety of molecules across the plant cell membrane to cause water soaking. Our discovery of AvrE/DspE inhibitors has significant implications in agriculture (provisional patent: DU7813PROV). Likewise, our structural elucidation of NPR1 and its complex with the transcription factor TGA3, carried out in collaboration with Dr. Xinnian Dong (Duke Biology), has profoundly transformed our knowledge about NPR1-mediated SAR in plants and has also led to the successful genetic engineering of plants with enhanced SAR and no loss of fitness (provisional patent: DU8135PROV).

• Structural basis of NPR1 in activating plant immunity. Kumar S†, Zavaliev R†, Wu Q†, Zhou Y, Cheng J, Dillard L, Powers J, Withers J, Zhao J, Guan Z, Borgnia MJ, Bartesaghi A, Dong X*, Zhou P*. Nature 2022; 605(7910): 561-566. (†equal contributions; *co-correspondence)
• Bacterial pathogens deliver water- and solute-permeable channels to plant cells. Nomura K†, Andreazza F†, Cheng J†, Dong K*, Zhou P* & He SY*. Nature. 2023; 621(7979):586-591. (†equal contributions; *co-correspondence)
3. Translesion DNA synthesis (TLS) and cancer therapy
Translesion DNA synthesis was initially discovered as a specialized DNA damage tolerance pathway to bypass DNA lesions during genomic replication. Recent studies have revealed an increasingly complex role of TLS in a variety of DNA damage response processes in cells. Our biochemical and structural elucidation of the Rev1-Pol -Rev3/7 quaternary complex (JBC 2012), in particular, has established the existence of a translesionsome (also known as “mutasome”) complex that carries insertion and extension activities in a single macromolecular machinery during TLS. This work has reshaped the established paradigm in the field, where insertion and extension activities were previously thought to function in distinct molecular complexes mediated by PCNA. Our lab further discovered a small molecule inhibitor, JH-RE-06 that disrupts the formation of the translesionsome complex and sensitizes tumor cells to chemotherapeutic agents in vitro and in vivo (Cell 2019). This compound, JH-RE-06, has now been widely used as a molecular probe to investigate the role of TLS in a variety of DNA repair processes.
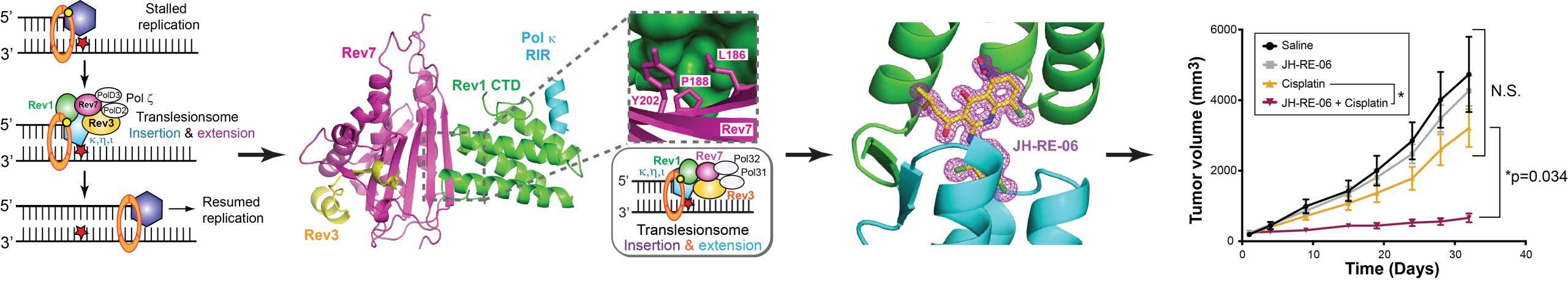
• A Small Molecule Targeting Mutagenic Translesion Synthesis Improves Chemotherapy. Wojtaszek JL, Chatterjee N, Najeeb J, Ramos A, Lee M, Bian K, Xue JY, Fenton BA, Park H, Li D, Hemann MT*, Hong J*, Walker GC*, and Zhou P*. Cell 2019; 178: 152-159. (PMC6644000; *co-correspondence)

4. Metabolic and glycosylation enzymes
Despite their significant roles in biology, the mechanism and function of many metabolic and glycosylation enzymes are not fully understood. In collaboration with Dr. Jen-Tsan Ashley Chi (Duke MGM), our lab elucidated the structure and mechanism of the NADPH phosphatase activity of human MESH1/HDDC3 (Nature Metabolism 2020). This work resolved a decade of debate about the biochemical and biological function of this enzyme, as MESH1 was initially suggested to hydrolyze ppGpp, a bacterial alarmone, except that ppGpp is largely undetectable in mammalian cells. Instead, our work showed that MESH1 functions as a NADPH hydrolase and regulates ferroptosis in mammalian cells. In another example, SPINDLY (SPY) in Arabidopsis thaliana was annotated as an O-linked-N-acetylglucosamine (GlcNAc) transferase (OGT) based on its sequence similarity to the human OGTs. However, our detailed biochemical and structural analysis, carried out in collaboration with Dr. Tai-Ping Sun (Duke Biology) and Dr. Alberto Bartesaghi (Duke Biochemistry and Computer Science), has provided the first concrete biochemical and structural evidence to establish SPY as a novel nucleocytoplasmic protein O-fucosyltransferase (POFUT) that regulates diverse developmental processes in plants. (Nat Chem Biol. 2017; Nat Commun. 2023)
• MESH1 is a cytosolic NADPH phosphatase that regulates ferroptosis. Ding CC, Rose J, Sun T, Wu J, Chen PH, Lin CC, Yang WH, Chen KY, Lee H, Xu E, Tian S, Akinwuntan J, Zhao J, Guan Z, Zhou P*, Chi JT*. Nat Metab. 2020 Mar;2(3):270-277. (*co-correspondence)
• Structure and dynamics of the Arabidopsis O-fucosyltransferase SPINDLY. Kumar S, Wang Y, Zhou Y, Dillard L, Li FW, Sciandra CA, Sui N, Zentella R, Zahn E, Shabanowitz J, Hunt DF, Borgnia MJ, Bartesaghi A*, Sun TP*, Zhou P*. Nat Commun. 2023; 14(1): 1538. (*co-correspondence)
5. Fast NMR Methodology
Dr. Zhou is a leading contributor to the development of the fast NMR methodology based on sparse, non-uniform sampling in the time domain followed by high-quality spectral reconstruction in the frequency domain. Such a technique accelerates NMR data acquisition by 10-100 folds compared with the conventional Nyquist sampling method without sacrificing spectral quality. This methodology has been adopted by commercial spectrometer vendors and has gained wide acceptance in the biomolecular NMR community with the arrival of ultra-high-field spectrometers exceeding 1 GHz.
• Coggins BE, Zhou P. High resolution 4-D spectroscopy with sparse concentric shell sampling and FFT-CLEAN. J Biomol NMR. 2008; 42(4):225-39. (PMC2680427)
• Wu Q, Coggins BE, Zhou P. Unbiased measurements of reconstruction fidelity of sparsely sampled magnetic resonance spectra. Nature Commun. 2016; 7: 12281. (PMC4974455)
