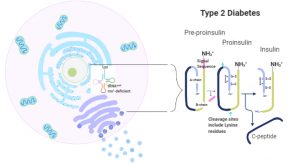
Type 2 Diabetes (T2D) is a condition characterized by insulin resistance and impaired insulin secretion, affecting over 30 million people, and is the seventh leading cause of death in the United States.
Genome-wide association studies have found that people with single nucleotide polymorphisms (SNPs) within intron 5 of cdkal1, a gene associated with insulin translation and processing, have an increased risk of developing T2D.
In pancreatic β-cells, cdkal1 encodes a tRNA-methylthiotransferase enzyme that modifies an already modified adenosine-37, N6-threonycarbamoyladenosine (t6A37), in tRNALys3, allowing the accurate reading of both lysine codons, AAA and AGG. Without this modification, tRNALys3 becomes inefficient and the frequency of mutations increases, preventing mature insulin from properly forming.
We are interested in investigating how the presence of SNPs within an intronic site of Cdkal1 could increase the risk of developing T2D. Without a functional Cdkal1 enzyme, a significant proportion of the population of tRNALys3 molecules may not be modified, which could be affecting how Cdkal1 mRNA and protein are produced.
We are currently studying the biophysical and enzymatic properties of the Cdkal1 protein, none of which has been done before.
For patients with cdkal1-associated diabetes, this lack of modified tRNAs, leading to mistranslated lysine codons, could be causing the disease. Although studies have shown that a lack of the cdkal1 gene reduces insulin production and impairs β-cell function, further investigation on how and which SNPs affect transcription of cdkal1 will help us understand the role of tRNA modification enzymes in T2D risk.