Glossary:
Promoter: A short sequence of DNA located just before the transcription start site for a gene, which is responsible for initiating the transcription of a gene, generally by recruiting RNA polymerase, the molecule responsible for DNA transcription.
Transcription factor: A protein that binds to specific sequences of DNA to control the rate of transcription, either decreasing or increasing it.
Enhancer: Similar to promoters, enhancers are involved in triggering the process of transcription. Enhancers are located within 100 kilobases up and downstream of the gene promoter, and are generally binding sites for transcription factors.
PV interneurons: One of three types of inhibitory, GABAergic interneurons (the others being VIP and SST), which can be identified by the expression of parvalbumin.
UCSC genome browser: A website curated by the University of California, Santa Cruz, that allows access to the genome sequence data of various vertebrates and invertebrates, including human, mouse, worm and fruitfly. Various annotations are available to provide visualization of various genomic factors along with the genes.
Adeno-associated virus: A common vector for genes that can hold up to 5 kilobases of genetic information.
Assay for Transposase-Accessible Chromatin with high throughput Sequencing (ATAC-seq): A technique that uses transposase Tn5 to measure areas of chromatin with low nucleosome density, or in other words, areas of accessible chromatin where transcription factors could bind.

The overall goal of my project this summer is to attempt to create a virus that can trigger cell type specific expression of genes. Previous methods to create such a tool have tried to use promoter sequences to achieve cell type specific gene expression, however these tools were not able to generate gene expression strictly in the targeted cell type. So, my project is to identify a sequence motif that is common to PV- specific enhancers, which can be included in the virus, along with a specific gene, to trigger cell type specific expression of that gene, instead of promoters. Although there is still a chance that this technology will fail, it is possible that enhancers will provide more cell specific gene expression. Promoters are more commonly regulated among cell types, whereas enhancers are likely more involved in cell type specific gene expression, based on differential epigenetic signatures.
The initial steps of this project involved identifying potential enhancer regions that are specific to PV cells. This process began by finding genes that are differentially expressed between PV and VIP neurons, as well as PV and excitatory neurons, using the data published by Mo. Once these genes had been identified, I used the UCSC genome browser, with the ATAC-seq data, also published by Mo, for the three cell types, to look for peaks that appear only in the PV cells, within 100 kb of differentially expressed genes. This part was slightly tedious because I had to search for the peaks by hand. Once the peaks had been identified, I converted the peak data to sequence data and entered it for motif analysis in the MEME-ChIP software, which identified novel and known transcription factor motifs. Once these motifs have been identified, they will be tested in vitro to ensure that the sequences are truly promoters, by the production of eRNA. Once that has been confirmed, we will begin to build the viruses and test their effectiveness as a tool to trigger cell specific gene expression.
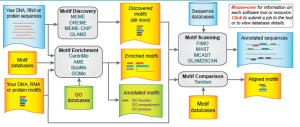
The flow of the motif finding software, which is based on statistical methods, and then compares the motifs found to known transcription factor binding sites.
