What is SEM model:
SEM is a combination of factor analysis and multiple regression
It also goes by the aliases “causal modeling” and “analysis of covariance structure”
Special cases of SEM include confirmatory factor analysis and path analysis
The SEM can be divided into two parts. The measurement model is the part which relates measured variables to latent variables. The structural model is the part that relates latent variables to one another.
The path analysis, which is SEM with no latent variables. In other words, path analysis is SEM with a structural model, but no measurement model. A structural equation modeling (SEM) has only single indicators are employed for each of the variables in the causal model.
SEM model in genetics study, I am collecting some lecture notes on SEM.
Karl Wuensch from ECU
David Kenny
Dr. William Revelle from North Western University
Dr. Brannick from USF has a good post on SEM vs. Path Analysis
David Kenny presents a very thorough explanation of using SEM
A very handy tutorial on SEM
It turns out that Dr. William Revelle from North Western University has a class Psychology 454 syllabus with detail R and R package for SEM.
In one of the lecture, he has a Lecture Note on “Latent Variable Modeling”.
An R package lavaan was created for Latent Variable Analysis and it works perfectly fine for an SEM model as shown in the following diagram.
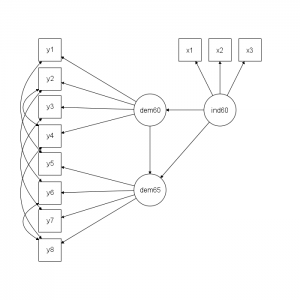
model <- '
# latent variables
ind60 =~ x1 + x2 + x3
dem60 =~ y1 + y2 + y3 + y4
dem65 =~ y5 + y6 + y7 + y8
# regressions
dem60 ~ ind60
dem65 ~ ind60 + dem60
# residual covariances
y1 ~~ y5
y2 ~~ y4 + y6
y3 ~~ y7
y4 ~~ y8
y6 ~~ y8
'
fit <- sem(model,
data=PoliticalDemocracy)
summary(fit)
Sometimes, there could be data-related “NOT converged” error! It is apparently data-dependent case. But, my model
mod1 <- '
SOX17_lev =~ GATA2_lev + PGR_lev
IHH ~ SOX17_lev '
happens to be like this!!
There has been a lot of resource from USGS — Dr. James Grace, who provides basic to advanced features and applications of SEM in his research.

