Random Forest implemented in python.
Wiki explanation gives the basic.
A collection of Machine Learning methods
Author Archives: Jin Tong
My first cup of python
How to install package “sklearn.ensemble”
I am learning Python using kaggle titanic project, the first error I got was, how can I install a package.
Here, is my answer under MacOX
Protected: Learning the GWAS analysis
Protected: Using LNCS1000 for Steve Kleeberger’s lab — Library of Integrated Network-based Cellular Signatures
Using HT-Seq for RNAseq project
cuffdiff output
There are 14 columns in cuffdiff output
awk -F"\t" '{print $1"\t"$8"\t"$9"\t"$10}' gene_exp.diff | head
test_id value_1 value_2 log2(fold_change)
A2M-AS1|144571|locus1of1|1 0.0190991 0.0447137 1.22722
A2MP1|3|locus1of1|1 0 0 0
A2M|2|locus1of1|1 0.0261932 0.0171011 -0.615099
A3GALT2|127550|locus1of1|1 0.0287783 0 -1.79769e+308
A4GNT|51146|locus1of1|1 0 0 0
AA06|100506677|locus1of1|1 0 0 0
AACSP1|729522|locus1of1|1 0 0.00822783 1.79769e+308
AADACL2-AS1|101928142|locus1of1|2 0 0 0
AADACL2|344752|locus1of1|1 0 0 0
...
the log2(fold_change) was computed by log2(value_2/value_1)
where “value_1” comes from the first “bam file” and “value_2” comes from the second “bam file” as the following command
cuffdiff -o /somePath/ /somePath/transcripts.gtf value_1_firstBam.bam value_2_secondBam.bam
tophat handles pair-end reads that overlap
Here is an old link
If the pairs are overlapped, they could be merged
It seems that the alingers do NOT care about the overlap and align them anyway. For tophat, it even allows negative insert input. It might not be relevant too much with RNAseq, where the read will be counted once toward the quantification. But, it will matter if one wants to detect peaks esp. display peaks on the broswer.
Protected: Display RNASeq data or any NGS track data on the UCSC genome browser — as a custom track
Using rshiny
Step 1, I need to create a repository on my github central place
Step 2 RShiny tutorial
Some special RShiny note
To start shinyapp on the command line, use the following command
R --vanilla -e "shiny::runApp('.', port=8888, host='10.91.128.1')"
R --vanilla -e "shiny::runApp('.', port=8888, host='localhost')"
Then go to a browser to load the application. For some reason, it fails to work with the follow 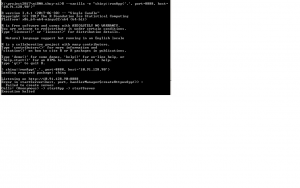 error message
error message
NGS data analysis protocols
Arraystar provides services
