
Electrophysiology

We use pressure-clamp (stretch) and cell-indentation (poke) electrophysiology to make precise measurements of many ion channel properties, including channel number, unitary conductance, ion selectivity, stimulus threshold and sensitivity, stimulus adaptation, and gating kinetics (activation, deactivation, inactivation, recovery from inactivation).
Super-resolution Microscopy
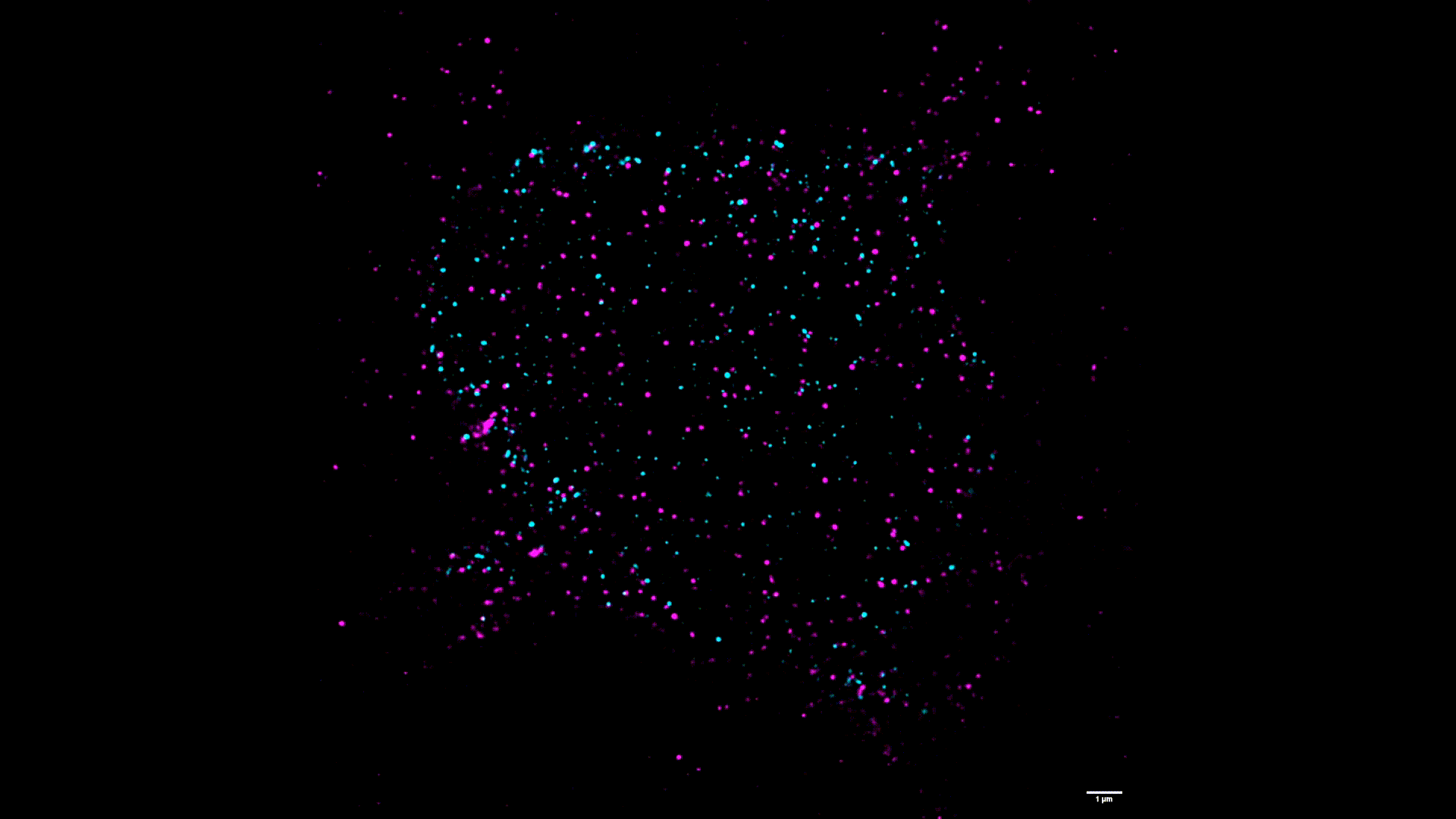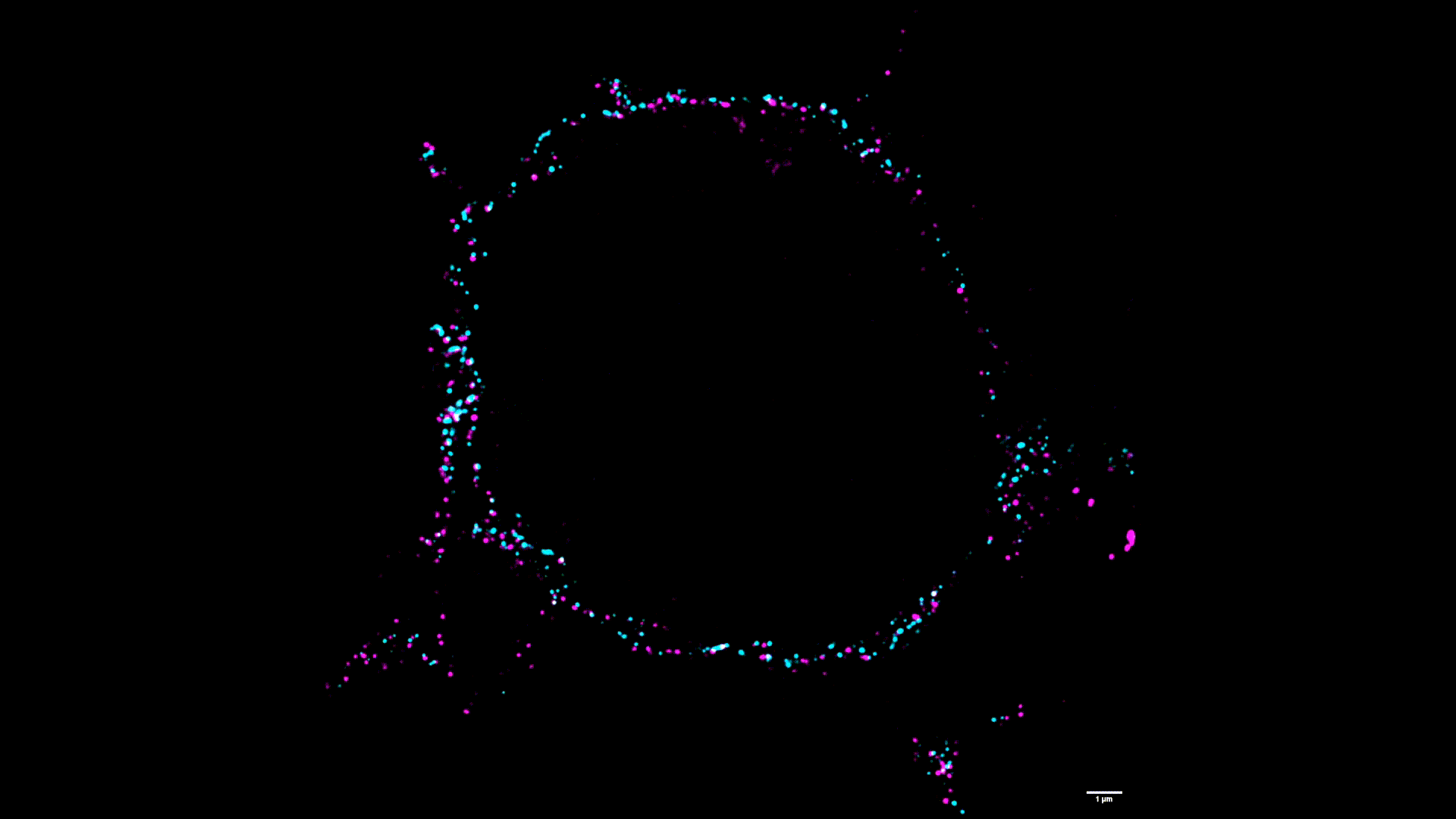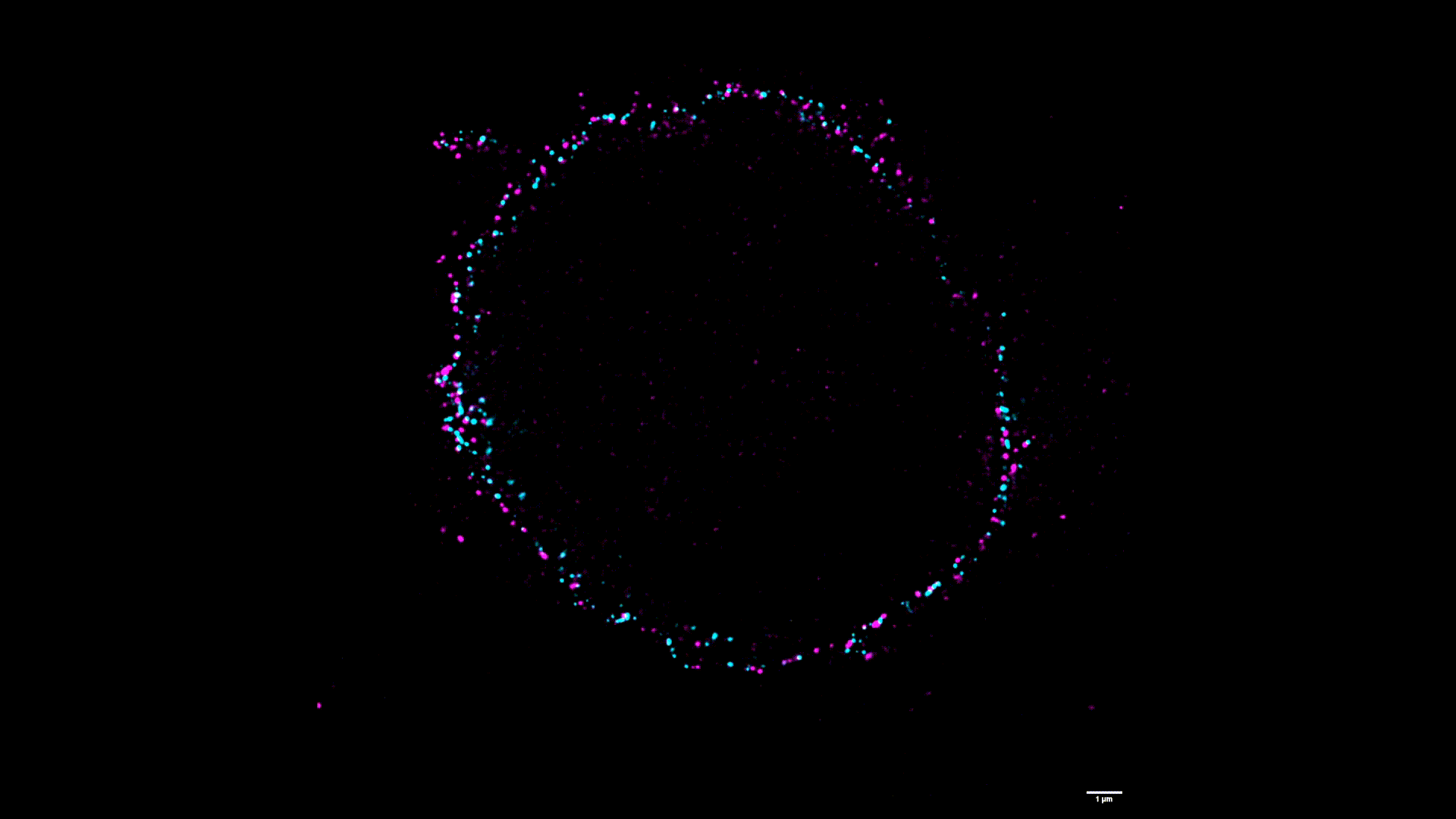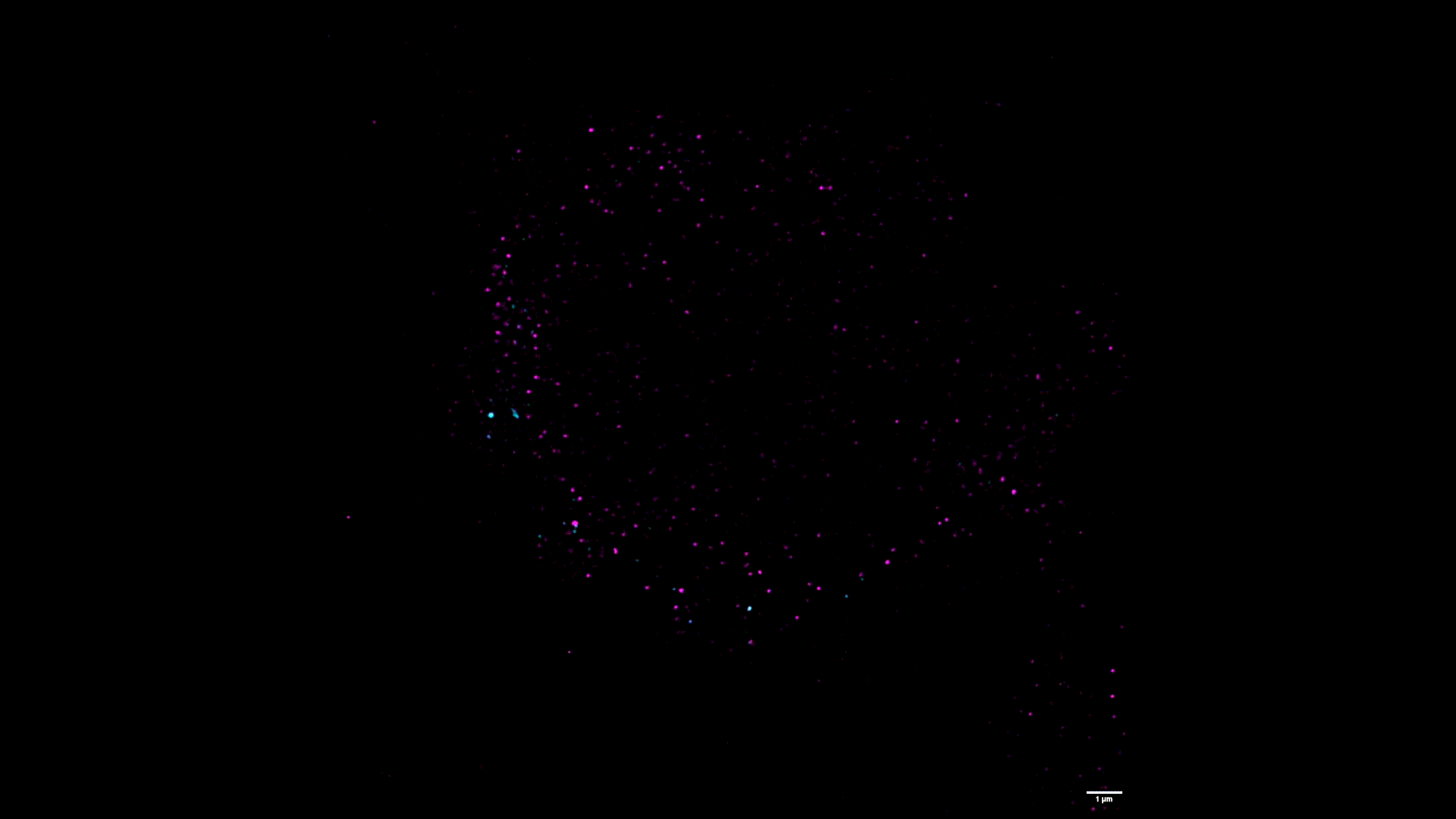
We use two-color stimulated emission-depletion (STED) microscopy to visualize the spatial distribution of force-gated ion channels with super-resolution precision (~80 nm).
Magnetic Force Application
By attaching superparamagnetic nanoparticles to specific domains of Piezo1 and exposing channels to a magnetic field while measuring channel activity electrophysiologically, we can probe their mechanical sensitivity with sub-molecular resolution.
Combined Atomic-Force Microscopy and Electrophysiology

We built an instrument that combines atomic force microscopy with patch-clamp electrophysiology to allow for quantitative mechanical stimulation and simultaneous detection of the evoked transduction current. With this tool, we are able to explore and precisely quantify how single cells convert the energy of mechanical compression into an electric signal.
Combined DIC Imaging and Electrophysiology
By combining high-magnification (400x) differential interference contrast (DIC) microscopy to image the geometry (radius) of a membrane patch containing force-gated ion channels, which allows calculating its membrane tension, with cell-attached pressure-clamp electrophysiology to apply mechanical stimulation (pressure) to the membrane patch and record the evoked response (current), we can measure tension-response curves with high precision.
Optically Guided Tension Clamp (OGTC)
Here, we further develop the approach of combining patch-clamp electrophysiology with differential interference contrast microscopy into a system that controls membrane tension in real time. The system uses machine learning object detection for millisecond analysis of membrane curvature and control of pipette pressure to produce a closed-loop membrane tension clamp.
Protein Structure and Function

Using structural information about force-gated ion channels we can introduce point-mutations, deletions, double-cysteine pairs for chemical crosslinking, and functional tags to probe molecular mechanisms.
Stochastic Simulations
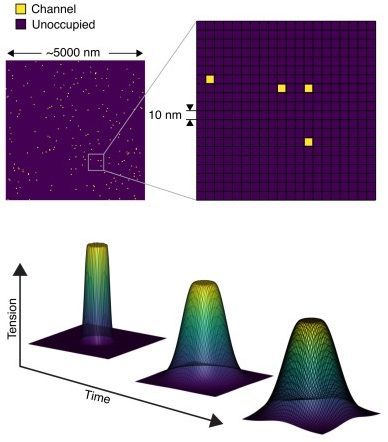
We can simulate the gating of thousands of individual, spatially randomly distributed Piezo1 channels (~100 channels/µm2) with a previously validated four-state Markov model in response to membrane tension diffusing in time and 2D space.