
Fractional anisotropy image, horizontal section through a 3D mouse brain volume

We study brain circuits using high resolution MRI.
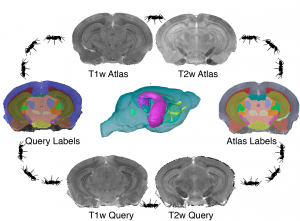
We develop multivariate image analysis pipelines for phenotyping mouse models of neurological and psychiatric conditions. We identify using statistical methods regions with different morphometric features (think volume as the simplest one), or microstructural properties.
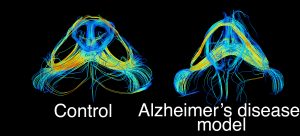
We examine tract based changes in mouse models of Alzheimer’s disease to identify vulnerable networks which can serve as targets for interventions.

We validate our translational, MRI based findings with alternative histological and microstructural markers. Here is an example of axonal cross section in a control mouse (left), versus a mouse model of Alzheimer’s disease.
We have used mouse models of Alzheimer’s disease developed by our collaborators at Duke (Dr Carol Colton, Duke Neurology), as well as outside Duke (Dr Joanna Jankowsky, Baylor).
Code developed by us, our collborators, and others – is available at GitHub. You can get an idea about what we do here: https://github.com/portokalh
The Visual Informatics Team at the Center for In Vivo Microscopy
Alexandra Badea, PhD, Robert J Anderson, PhD; James J Cook, BSc
With gracious help from Michele Wang, BSc candidate, Neuroscience, CS

Hi, this is a comment.
To get started with moderating, editing, and deleting comments, please visit the Comments screen in the dashboard.
Commenter avatars come from Gravatar.